Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
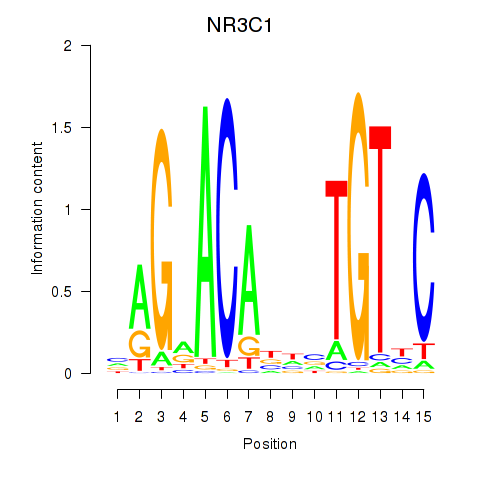
Results for NR3C1
Z-value: 0.95
Transcription factors associated with NR3C1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR3C1
|
ENSG00000113580.15 | NR3C1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR3C1 | hg38_v1_chr5_-_143403297_143403329 | 0.54 | 5.0e-03 | Click! |
Activity profile of NR3C1 motif
Sorted Z-values of NR3C1 motif
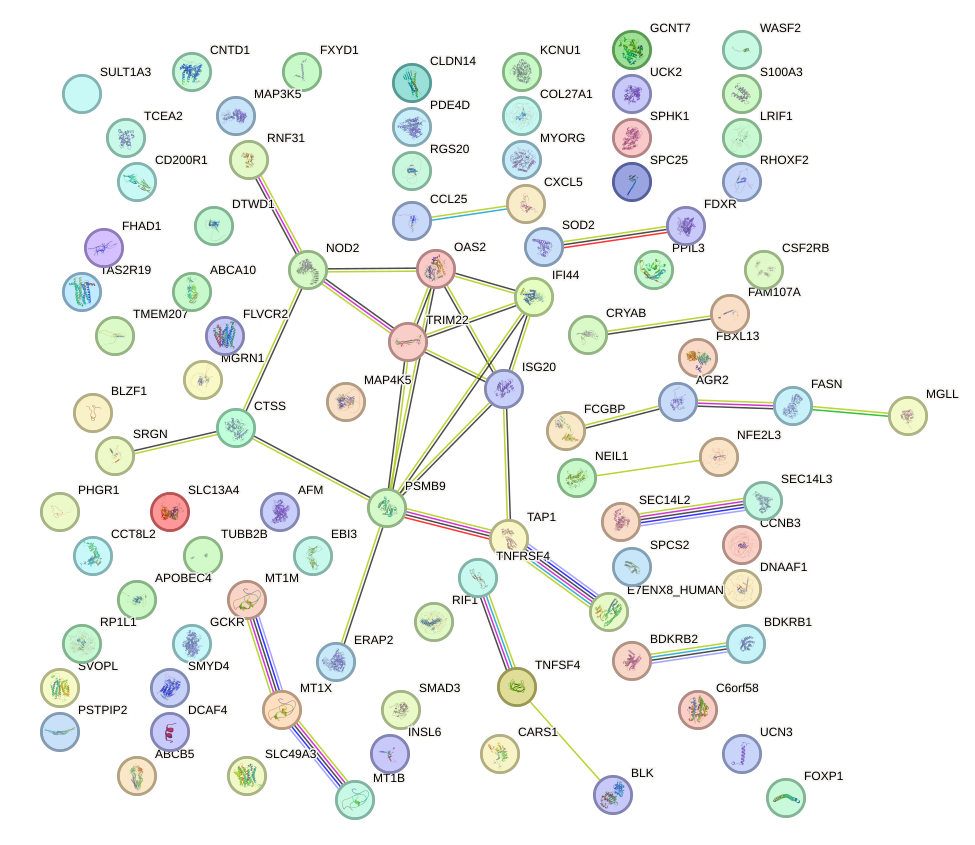
Network of associatons between targets according to the STRING database.
First level regulatory network of NR3C1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_88635626 | 2.50 |
ENST00000379224.10
|
ISG20
|
interferon stimulated exonuclease gene 20 |
| chr19_+_4229502 | 1.95 |
ENST00000221847.6
|
EBI3
|
Epstein-Barr virus induced 3 |
| chr6_+_127577168 | 1.61 |
ENST00000329722.8
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr9_+_114155526 | 1.46 |
ENST00000356083.8
|
COL27A1
|
collagen type XXVII alpha 1 chain |
| chr2_-_224982420 | 1.39 |
ENST00000645028.1
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr2_-_168913277 | 1.39 |
ENST00000451987.5
|
SPC25
|
SPC25 component of NDC80 kinetochore complex |
| chr4_-_687325 | 1.33 |
ENST00000503156.5
|
SLC49A3
|
solute carrier family 49 member 3 |
| chr3_-_128052166 | 1.28 |
ENST00000648300.1
|
MGLL
|
monoglyceride lipase |
| chr3_-_127822835 | 1.28 |
ENST00000453507.6
|
MGLL
|
monoglyceride lipase |
| chr1_-_1214146 | 1.16 |
ENST00000379236.4
|
TNFRSF4
|
TNF receptor superfamily member 4 |
| chr2_-_224947030 | 1.15 |
ENST00000409592.7
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr18_-_46072244 | 1.14 |
ENST00000589328.5
ENST00000409746.5 |
PSTPIP2
|
proline-serine-threonine phosphatase interacting protein 2 |
| chr1_-_173205543 | 1.12 |
ENST00000367718.5
|
TNFSF4
|
TNF superfamily member 4 |
| chr14_+_96204679 | 1.11 |
ENST00000542454.2
ENST00000539359.1 ENST00000554311.2 ENST00000553811.1 |
BDKRB2
ENSG00000258691.1
|
bradykinin receptor B2 novel protein |
| chr3_-_127822455 | 1.09 |
ENST00000265052.10
|
MGLL
|
monoglyceride lipase |
| chr1_-_150765735 | 0.99 |
ENST00000679898.1
ENST00000448301.7 ENST00000680664.1 ENST00000679512.1 ENST00000368985.8 ENST00000679582.1 |
CTSS
|
cathepsin S |
| chr6_-_32853813 | 0.98 |
ENST00000643049.2
|
TAP1
|
transporter 1, ATP binding cassette subfamily B member |
| chr16_+_56682461 | 0.98 |
ENST00000562939.1
ENST00000394485.5 ENST00000567563.1 |
MT1X
ENSG00000259827.1
|
metallothionein 1X novel transcript |
| chr5_+_96876480 | 0.94 |
ENST00000437043.8
ENST00000379904.8 |
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chrX_+_50204753 | 0.90 |
ENST00000376042.6
|
CCNB3
|
cyclin B3 |
| chr6_-_32853618 | 0.90 |
ENST00000354258.5
|
TAP1
|
transporter 1, ATP binding cassette subfamily B member |
| chr1_+_165827574 | 0.89 |
ENST00000367879.9
|
UCK2
|
uridine-cytidine kinase 2 |
| chr6_+_32854179 | 0.89 |
ENST00000374859.3
|
PSMB9
|
proteasome 20S subunit beta 9 |
| chr5_+_114056017 | 0.88 |
ENST00000512097.9
|
KCNN2
|
potassium calcium-activated channel subfamily N member 2 |
| chr1_+_165827786 | 0.84 |
ENST00000642653.1
|
UCK2
|
uridine-cytidine kinase 2 |
| chr1_-_153549238 | 0.84 |
ENST00000368713.8
|
S100A3
|
S100 calcium binding protein A3 |
| chr4_-_73998669 | 0.84 |
ENST00000296027.5
|
CXCL5
|
C-X-C motif chemokine ligand 5 |
| chr8_+_53880894 | 0.82 |
ENST00000276500.4
|
RGS20
|
regulator of G protein signaling 20 |
| chr8_+_53880867 | 0.81 |
ENST00000522225.5
|
RGS20
|
regulator of G protein signaling 20 |
| chr1_+_15345084 | 0.80 |
ENST00000529606.5
|
FHAD1
|
forkhead associated phosphopeptide binding domain 1 |
| chr1_-_153549120 | 0.79 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr7_-_16833411 | 0.78 |
ENST00000412973.1
|
AGR2
|
anterior gradient 2, protein disulphide isomerase family member |
| chr12_-_11022620 | 0.78 |
ENST00000390673.2
|
TAS2R19
|
taste 2 receptor member 19 |
| chr16_+_4624811 | 0.77 |
ENST00000415496.5
ENST00000262370.12 ENST00000587747.5 ENST00000399577.9 ENST00000588994.5 ENST00000586183.5 |
MGRN1
|
mahogunin ring finger 1 |
| chr20_+_64063481 | 0.76 |
ENST00000415602.5
|
TCEA2
|
transcription elongation factor A2 |
| chr17_-_69150062 | 0.75 |
ENST00000522787.5
ENST00000521538.5 |
ABCA10
|
ATP binding cassette subfamily A member 10 |
| chr16_+_56632651 | 0.74 |
ENST00000379818.4
ENST00000570233.1 |
MT1M
|
metallothionein 1M |
| chr5_-_59216826 | 0.74 |
ENST00000638939.1
|
PDE4D
|
phosphodiesterase 4D |
| chr16_+_33827140 | 0.73 |
ENST00000562905.2
|
IGHV3OR16-13
|
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
| chr16_+_84176067 | 0.71 |
ENST00000564928.1
|
DNAAF1
|
dynein axonemal assembly factor 1 |
| chr6_-_32838727 | 0.71 |
ENST00000652259.1
ENST00000374897.4 ENST00000620123.4 ENST00000452392.2 |
TAP2
ENSG00000250264.1
|
transporter 2, ATP binding cassette subfamily B member novel protein, TAP2-HLA-DOB readthrough |
| chr6_-_27867581 | 0.70 |
ENST00000331442.5
|
H1-5
|
H1.5 linker histone, cluster member |
| chr11_-_60243103 | 0.68 |
ENST00000651255.1
|
MS4A4E
|
membrane spanning 4-domains A4E |
| chr12_+_112978562 | 0.68 |
ENST00000680122.1
|
OAS2
|
2'-5'-oligoadenylate synthetase 2 |
| chr17_+_42798779 | 0.67 |
ENST00000585355.5
|
CNTD1
|
cyclin N-terminal domain containing 1 |
| chr2_-_89213917 | 0.67 |
ENST00000498435.1
|
IGKV1-27
|
immunoglobulin kappa variable 1-27 |
| chr11_+_5689780 | 0.66 |
ENST00000379965.8
ENST00000454828.5 |
TRIM22
|
tripartite motif containing 22 |
| chr19_+_8052752 | 0.66 |
ENST00000315626.6
ENST00000253451.9 |
CCL25
|
C-C motif chemokine ligand 25 |
| chr19_+_14072754 | 0.64 |
ENST00000587086.2
|
MISP3
|
MISP family member 3 |
| chr6_-_159693228 | 0.64 |
ENST00000367054.6
ENST00000367055.8 ENST00000538183.7 ENST00000444946.6 ENST00000452684.2 |
SOD2
|
superoxide dismutase 2 |
| chr19_+_35139440 | 0.64 |
ENST00000455515.6
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr19_+_35139724 | 0.62 |
ENST00000588715.5
ENST00000588607.5 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr19_+_8053000 | 0.61 |
ENST00000390669.7
|
CCL25
|
C-C motif chemokine ligand 25 |
| chr16_+_56625775 | 0.60 |
ENST00000330439.7
ENST00000568293.1 |
MT1E
|
metallothionein 1E |
| chr19_+_35140022 | 0.60 |
ENST00000588081.5
ENST00000589121.1 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr2_+_27496830 | 0.59 |
ENST00000264717.7
|
GCKR
|
glucokinase regulator |
| chr6_+_32844108 | 0.59 |
ENST00000458296.2
ENST00000413039.6 ENST00000412095.1 ENST00000395330.5 |
PSMB8-AS1
PSMB9
|
PSMB8 antisense RNA 1 (head to head) proteasome 20S subunit beta 9 |
| chr3_-_112975018 | 0.59 |
ENST00000471858.5
ENST00000308611.8 ENST00000295863.4 |
CD200R1
|
CD200 receptor 1 |
| chr6_-_3227643 | 0.58 |
ENST00000259818.8
|
TUBB2B
|
tubulin beta 2B class IIb |
| chr1_-_183653307 | 0.58 |
ENST00000308641.6
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme catalytic polypeptide like 4 |
| chr3_-_190449782 | 0.58 |
ENST00000354905.3
|
TMEM207
|
transmembrane protein 207 |
| chr10_+_69088096 | 0.57 |
ENST00000242465.4
|
SRGN
|
serglycin |
| chr3_+_119146184 | 0.57 |
ENST00000460150.1
|
TEX55
|
testis expressed 55 |
| chr1_-_27490045 | 0.57 |
ENST00000536657.1
|
WASF2
|
WASP family member 2 |
| chr9_-_34372832 | 0.57 |
ENST00000379142.3
|
MYORG
|
myogenesis regulating glycosidase (putative) |
| chr1_-_173207322 | 0.56 |
ENST00000281834.4
|
TNFSF4
|
TNF superfamily member 4 |
| chr16_+_84175933 | 0.55 |
ENST00000569735.1
|
DNAAF1
|
dynein axonemal assembly factor 1 |
| chr8_-_10655137 | 0.55 |
ENST00000382483.4
|
RP1L1
|
RP1 like 1 |
| chr16_+_30201057 | 0.55 |
ENST00000569485.5
|
SULT1A3
|
sulfotransferase family 1A member 3 |
| chr20_-_56525925 | 0.53 |
ENST00000243913.8
|
GCNT7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr17_+_76384601 | 0.53 |
ENST00000592299.6
ENST00000590959.5 ENST00000323374.8 |
SPHK1
|
sphingosine kinase 1 |
| chr15_+_67125707 | 0.53 |
ENST00000540846.6
|
SMAD3
|
SMAD family member 3 |
| chr8_+_11494367 | 0.52 |
ENST00000259089.9
ENST00000529894.1 |
BLK
|
BLK proto-oncogene, Src family tyrosine kinase |
| chr21_-_36480117 | 0.51 |
ENST00000399135.6
|
CLDN14
|
claudin 14 |
| chr22_+_36913620 | 0.51 |
ENST00000403662.8
ENST00000262825.9 |
CSF2RB
|
colony stimulating factor 2 receptor subunit beta |
| chr6_-_136792466 | 0.51 |
ENST00000359015.5
|
MAP3K5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr21_-_36480060 | 0.51 |
ENST00000399137.5
|
CLDN14
|
claudin 14 |
| chr10_+_5364955 | 0.51 |
ENST00000380433.5
|
UCN3
|
urocortin 3 |
| chr7_+_20615653 | 0.50 |
ENST00000404938.7
|
ABCB5
|
ATP binding cassette subfamily B member 5 |
| chr16_+_56651885 | 0.50 |
ENST00000334346.3
ENST00000562399.1 |
MT1B
|
metallothionein 1B |
| chr17_-_74868616 | 0.50 |
ENST00000579893.1
ENST00000544854.5 |
FDXR
|
ferredoxin reductase |
| chr14_+_24147474 | 0.49 |
ENST00000324103.11
ENST00000559260.5 |
RNF31
|
ring finger protein 31 |
| chr3_-_58577367 | 0.49 |
ENST00000464064.5
ENST00000360997.7 |
FAM107A
|
family with sequence similarity 107 member A |
| chr7_-_135728177 | 0.49 |
ENST00000682651.1
ENST00000354042.8 |
SLC13A4
|
solute carrier family 13 member 4 |
| chr4_+_73481737 | 0.49 |
ENST00000226355.5
|
AFM
|
afamin |
| chr15_+_40351026 | 0.48 |
ENST00000448599.2
|
PHGR1
|
proline, histidine and glycine rich 1 |
| chr1_+_78649818 | 0.47 |
ENST00000370747.9
ENST00000438486.1 |
IFI44
|
interferon induced protein 44 |
| chr3_-_58577648 | 0.47 |
ENST00000394481.5
|
FAM107A
|
family with sequence similarity 107 member A |
| chr7_-_103074816 | 0.46 |
ENST00000379305.7
ENST00000379308.7 |
FBXL13
|
F-box and leucine rich repeat protein 13 |
| chr17_+_76385256 | 0.45 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr1_+_169368175 | 0.45 |
ENST00000367808.8
ENST00000426663.1 |
BLZF1
|
basic leucine zipper nuclear factor 1 |
| chr15_+_49621004 | 0.44 |
ENST00000403028.8
ENST00000558653.5 ENST00000559164.5 ENST00000560632.5 ENST00000559405.5 ENST00000251250.7 |
DTWD1
|
DTW domain containing 1 |
| chr9_-_131709721 | 0.44 |
ENST00000372190.7
|
RAPGEF1
|
Rap guanine nucleotide exchange factor 1 |
| chr7_-_138627444 | 0.43 |
ENST00000463557.1
|
SVOPL
|
SVOP like |
| chr22_-_16592810 | 0.43 |
ENST00000359963.4
|
CCT8L2
|
chaperonin containing TCP1 subunit 8 like 2 |
| chr8_+_36784324 | 0.43 |
ENST00000523973.5
ENST00000399881.8 |
KCNU1
|
potassium calcium-activated channel subfamily U member 1 |
| chr15_+_75347727 | 0.43 |
ENST00000565051.5
ENST00000564257.1 ENST00000567005.1 |
NEIL1
|
nei like DNA glycosylase 1 |
| chr11_-_3057386 | 0.43 |
ENST00000529772.5
ENST00000278224.13 ENST00000380525.9 |
CARS1
|
cysteinyl-tRNA synthetase 1 |
| chrX_+_120158600 | 0.42 |
ENST00000371388.5
|
RHOXF2
|
Rhox homeobox family member 2 |
| chr2_+_151409878 | 0.42 |
ENST00000453091.6
ENST00000428287.6 ENST00000444746.7 ENST00000243326.9 ENST00000414861.6 |
RIF1
|
replication timing regulatory factor 1 |
| chr3_-_71130963 | 0.41 |
ENST00000649695.2
|
FOXP1
|
forkhead box P1 |
| chr2_-_2326378 | 0.41 |
ENST00000647618.1
|
MYT1L
|
myelin transcription factor 1 like |
| chr3_-_71130892 | 0.41 |
ENST00000491238.7
ENST00000674446.1 |
FOXP1
|
forkhead box P1 |
| chr14_+_75632610 | 0.41 |
ENST00000555027.1
|
FLVCR2
|
FLVCR heme transporter 2 |
| chr16_+_50696999 | 0.41 |
ENST00000300589.6
|
NOD2
|
nucleotide binding oligomerization domain containing 2 |
| chr17_-_1829818 | 0.41 |
ENST00000305513.12
|
SMYD4
|
SET and MYND domain containing 4 |
| chr5_-_59586393 | 0.41 |
ENST00000505453.1
ENST00000360047.9 |
PDE4D
|
phosphodiesterase 4D |
| chr11_-_111912871 | 0.40 |
ENST00000528628.5
|
CRYAB
|
crystallin alpha B |
| chr7_+_26152188 | 0.40 |
ENST00000056233.4
|
NFE2L3
|
nuclear factor, erythroid 2 like 3 |
| chr19_-_39934626 | 0.40 |
ENST00000616721.6
|
FCGBP
|
Fc fragment of IgG binding protein |
| chrX_+_9463272 | 0.40 |
ENST00000407597.7
ENST00000380961.5 ENST00000424279.6 |
TBL1X
|
transducin beta like 1 X-linked |
| chr14_+_72926377 | 0.40 |
ENST00000353777.7
ENST00000358377.7 ENST00000394234.6 ENST00000509153.5 ENST00000555042.5 |
DCAF4
|
DDB1 and CUL4 associated factor 4 |
| chr10_+_88990736 | 0.40 |
ENST00000357339.6
ENST00000652046.1 ENST00000355279.2 |
FAS
|
Fas cell surface death receptor |
| chr15_-_78944985 | 0.40 |
ENST00000615999.5
ENST00000677789.1 ENST00000676880.1 ENST00000677936.1 ENST00000220166.10 ENST00000677810.1 ENST00000678644.1 ENST00000677534.1 ENST00000677316.1 |
CTSH
|
cathepsin H |
| chr5_+_68292562 | 0.40 |
ENST00000523872.1
|
PIK3R1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr4_+_80197493 | 0.39 |
ENST00000415738.3
|
PRDM8
|
PR/SET domain 8 |
| chrX_+_3066833 | 0.39 |
ENST00000359361.2
|
ARSF
|
arylsulfatase F |
| chr14_-_52791597 | 0.39 |
ENST00000216410.8
ENST00000557604.1 |
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chrX_-_153446051 | 0.39 |
ENST00000370231.3
|
TREX2
|
three prime repair exonuclease 2 |
| chr19_-_52962837 | 0.39 |
ENST00000391786.6
ENST00000357666.8 ENST00000438970.6 ENST00000270457.8 ENST00000535506.5 ENST00000444460.7 ENST00000457013.6 |
ZNF816
|
zinc finger protein 816 |
| chr12_+_104303726 | 0.38 |
ENST00000527879.2
|
EID3
|
EP300 interacting inhibitor of differentiation 3 |
| chr8_-_22156789 | 0.38 |
ENST00000306317.7
|
LGI3
|
leucine rich repeat LGI family member 3 |
| chr11_-_34511710 | 0.38 |
ENST00000620316.4
ENST00000312319.6 |
ELF5
|
E74 like ETS transcription factor 5 |
| chr10_+_113679839 | 0.38 |
ENST00000369318.8
ENST00000369315.5 |
CASP7
|
caspase 7 |
| chr6_-_27312258 | 0.38 |
ENST00000444565.2
|
POM121L2
|
POM121 transmembrane nucleoporin like 2 |
| chr8_-_22156741 | 0.37 |
ENST00000424267.6
|
LGI3
|
leucine rich repeat LGI family member 3 |
| chr16_+_56608577 | 0.37 |
ENST00000245185.6
ENST00000561491.1 |
MT2A
|
metallothionein 2A |
| chr6_-_17987463 | 0.37 |
ENST00000378814.9
ENST00000636847.1 ENST00000378843.6 ENST00000378826.6 ENST00000502704.2 ENST00000259711.11 |
KIF13A
|
kinesin family member 13A |
| chr13_+_87671354 | 0.37 |
ENST00000683689.1
|
SLITRK5
|
SLIT and NTRK like family member 5 |
| chr18_+_34818436 | 0.37 |
ENST00000599844.5
ENST00000679731.1 |
DTNA
|
dystrobrevin alpha |
| chr1_+_109539865 | 0.36 |
ENST00000618721.4
ENST00000527748.5 ENST00000616874.1 |
GPR61
|
G protein-coupled receptor 61 |
| chr2_+_197705353 | 0.36 |
ENST00000282276.8
|
MARS2
|
methionyl-tRNA synthetase 2, mitochondrial |
| chrX_+_152914426 | 0.36 |
ENST00000318504.11
ENST00000449285.6 ENST00000539731.5 ENST00000535861.5 ENST00000370268.8 ENST00000370270.6 |
ZNF185
|
zinc finger protein 185 with LIM domain |
| chr2_+_85753984 | 0.36 |
ENST00000306279.4
|
ATOH8
|
atonal bHLH transcription factor 8 |
| chr3_+_119146159 | 0.35 |
ENST00000295622.6
|
TEX55
|
testis expressed 55 |
| chr14_+_93333210 | 0.35 |
ENST00000256339.8
|
UNC79
|
unc-79 homolog, NALCN channel complex subunit |
| chr2_-_89297785 | 0.35 |
ENST00000465170.1
|
IGKV1-37
|
immunoglobulin kappa variable 1-37 (non-functional) |
| chr6_-_154510675 | 0.35 |
ENST00000607772.6
|
CNKSR3
|
CNKSR family member 3 |
| chr2_-_2326210 | 0.34 |
ENST00000647755.1
|
MYT1L
|
myelin transcription factor 1 like |
| chr15_+_74788542 | 0.34 |
ENST00000567571.5
|
CSK
|
C-terminal Src kinase |
| chr13_+_76952467 | 0.34 |
ENST00000449753.2
|
ACOD1
|
aconitate decarboxylase 1 |
| chr2_+_17878637 | 0.34 |
ENST00000304101.9
|
KCNS3
|
potassium voltage-gated channel modifier subfamily S member 3 |
| chr3_+_130931893 | 0.34 |
ENST00000504612.5
|
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr16_+_3065380 | 0.34 |
ENST00000551122.5
ENST00000548807.5 ENST00000528163.6 |
IL32
|
interleukin 32 |
| chr6_+_31670379 | 0.34 |
ENST00000409525.1
|
LY6G5B
|
lymphocyte antigen 6 family member G5B |
| chr3_+_156674579 | 0.34 |
ENST00000295924.12
|
TIPARP
|
TCDD inducible poly(ADP-ribose) polymerase |
| chr4_-_78939352 | 0.33 |
ENST00000512733.5
|
PAQR3
|
progestin and adipoQ receptor family member 3 |
| chr6_-_89217339 | 0.33 |
ENST00000454853.7
|
GABRR1
|
gamma-aminobutyric acid type A receptor subunit rho1 |
| chr6_+_69866546 | 0.33 |
ENST00000620364.5
|
COL19A1
|
collagen type XIX alpha 1 chain |
| chr22_-_26479664 | 0.33 |
ENST00000402105.7
|
HPS4
|
HPS4 biogenesis of lysosomal organelles complex 3 subunit 2 |
| chr1_-_155300979 | 0.33 |
ENST00000392414.7
|
PKLR
|
pyruvate kinase L/R |
| chr1_-_173824322 | 0.33 |
ENST00000356198.6
|
CENPL
|
centromere protein L |
| chr3_-_69052309 | 0.32 |
ENST00000398559.7
ENST00000646708.1 ENST00000646304.1 |
TMF1
|
TATA element modulatory factor 1 |
| chr7_+_134646845 | 0.32 |
ENST00000344924.8
|
BPGM
|
bisphosphoglycerate mutase |
| chr11_+_107591077 | 0.32 |
ENST00000531234.5
ENST00000265840.12 |
ELMOD1
|
ELMO domain containing 1 |
| chr18_+_24113341 | 0.32 |
ENST00000540918.2
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr4_+_37891060 | 0.32 |
ENST00000261439.9
ENST00000508802.5 ENST00000402522.1 |
TBC1D1
|
TBC1 domain family member 1 |
| chr1_-_205850125 | 0.32 |
ENST00000367136.5
|
PM20D1
|
peptidase M20 domain containing 1 |
| chr22_+_50738198 | 0.31 |
ENST00000216139.10
ENST00000529621.1 |
ACR
|
acrosin |
| chr22_+_24607638 | 0.31 |
ENST00000432867.5
|
GGT1
|
gamma-glutamyltransferase 1 |
| chr18_+_74499861 | 0.31 |
ENST00000324301.12
|
CNDP2
|
carnosine dipeptidase 2 |
| chr7_+_134646798 | 0.31 |
ENST00000418040.5
ENST00000393132.2 |
BPGM
|
bisphosphoglycerate mutase |
| chr11_-_9265078 | 0.31 |
ENST00000530044.5
ENST00000679568.1 ENST00000680294.1 ENST00000681203.1 |
DENND5A
|
DENN domain containing 5A |
| chr11_-_47352693 | 0.31 |
ENST00000256993.8
ENST00000399249.6 ENST00000545968.6 |
MYBPC3
|
myosin binding protein C3 |
| chr17_-_3063607 | 0.31 |
ENST00000575751.1
|
OR1D5
|
olfactory receptor family 1 subfamily D member 5 |
| chr5_+_134114673 | 0.31 |
ENST00000342854.10
ENST00000395029.5 |
TCF7
|
transcription factor 7 |
| chr1_+_15617415 | 0.31 |
ENST00000480945.6
|
DDI2
|
DNA damage inducible 1 homolog 2 |
| chrX_-_23743201 | 0.30 |
ENST00000492081.1
ENST00000379303.10 ENST00000336430.11 |
ACOT9
|
acyl-CoA thioesterase 9 |
| chr14_-_52069039 | 0.30 |
ENST00000216286.10
|
NID2
|
nidogen 2 |
| chr12_-_6342020 | 0.30 |
ENST00000540022.5
ENST00000536194.1 |
TNFRSF1A
|
TNF receptor superfamily member 1A |
| chr4_+_69280472 | 0.30 |
ENST00000335568.10
ENST00000511240.1 |
UGT2B28
|
UDP glucuronosyltransferase family 2 member B28 |
| chr17_+_75979211 | 0.30 |
ENST00000397640.6
ENST00000588202.5 ENST00000590676.1 ENST00000586891.1 |
TEN1
|
TEN1 subunit of CST complex |
| chr1_-_146144804 | 0.30 |
ENST00000583866.9
ENST00000617010.2 |
NBPF10
|
NBPF member 10 |
| chr3_+_9902619 | 0.30 |
ENST00000421412.5
|
IL17RE
|
interleukin 17 receptor E |
| chr13_-_33350619 | 0.30 |
ENST00000439831.1
ENST00000567873.1 |
STARD13
|
StAR related lipid transfer domain containing 13 |
| chr11_+_34642468 | 0.30 |
ENST00000527935.1
|
EHF
|
ETS homologous factor |
| chr3_-_11582330 | 0.30 |
ENST00000451674.6
|
VGLL4
|
vestigial like family member 4 |
| chr9_+_122159886 | 0.30 |
ENST00000373764.8
ENST00000536616.5 |
MORN5
|
MORN repeat containing 5 |
| chr22_+_22357739 | 0.30 |
ENST00000390294.2
|
IGLV1-47
|
immunoglobulin lambda variable 1-47 |
| chr10_+_113679523 | 0.29 |
ENST00000345633.8
ENST00000614447.4 ENST00000369321.6 |
CASP7
|
caspase 7 |
| chr2_+_90154073 | 0.29 |
ENST00000611391.1
|
IGKV1D-13
|
immunoglobulin kappa variable 1D-13 |
| chr11_+_308408 | 0.29 |
ENST00000399815.2
|
ENSG00000288681.1
|
novel protein |
| chr1_-_222712428 | 0.29 |
ENST00000355727.3
ENST00000340020.11 |
AIDA
|
axin interactor, dorsalization associated |
| chr12_+_100794769 | 0.29 |
ENST00000392977.8
ENST00000546991.1 ENST00000392979.7 |
ANO4
|
anoctamin 4 |
| chr1_-_160523204 | 0.29 |
ENST00000368055.1
ENST00000368057.8 ENST00000368059.7 |
SLAMF6
|
SLAM family member 6 |
| chr14_-_106235582 | 0.29 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr2_+_89936859 | 0.29 |
ENST00000474213.1
|
IGKV2D-30
|
immunoglobulin kappa variable 2D-30 |
| chr4_-_155376797 | 0.29 |
ENST00000650955.1
ENST00000515654.5 |
MAP9
|
microtubule associated protein 9 |
| chr18_+_58341038 | 0.29 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr16_-_12803785 | 0.29 |
ENST00000433677.6
ENST00000261660.4 ENST00000381774.9 |
CPPED1
|
calcineurin like phosphoesterase domain containing 1 |
| chr17_+_59155726 | 0.29 |
ENST00000578777.5
ENST00000577457.1 ENST00000582995.5 ENST00000262293.9 ENST00000614081.1 |
PRR11
|
proline rich 11 |
| chr15_-_72197772 | 0.29 |
ENST00000309731.12
|
GRAMD2A
|
GRAM domain containing 2A |
| chr14_-_106811131 | 0.29 |
ENST00000424969.2
|
IGHV3-74
|
immunoglobulin heavy variable 3-74 |
| chr1_+_155613221 | 0.29 |
ENST00000462250.2
|
MSTO1
|
misato mitochondrial distribution and morphology regulator 1 |
| chr14_-_106130061 | 0.29 |
ENST00000390602.3
|
IGHV3-13
|
immunoglobulin heavy variable 3-13 |
| chr2_+_26034069 | 0.29 |
ENST00000264710.5
|
RAB10
|
RAB10, member RAS oncogene family |
| chr4_+_146638890 | 0.29 |
ENST00000281321.3
|
POU4F2
|
POU class 4 homeobox 2 |
| chr11_-_125778788 | 0.28 |
ENST00000436890.2
|
PATE2
|
prostate and testis expressed 2 |
| chr1_-_27366858 | 0.28 |
ENST00000374040.7
|
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr12_-_6342066 | 0.28 |
ENST00000162749.7
ENST00000440083.6 |
TNFRSF1A
|
TNF receptor superfamily member 1A |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.6 | 2.5 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.6 | 1.7 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.4 | 1.7 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.4 | 3.7 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 | 1.0 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.3 | 1.0 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.3 | 1.9 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.2 | 1.5 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.2 | 1.3 | GO:0036018 | cellular response to erythropoietin(GO:0036018) |
| 0.2 | 0.6 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.2 | 0.3 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.2 | 0.5 | GO:0038156 | interleukin-5-mediated signaling pathway(GO:0038043) interleukin-3-mediated signaling pathway(GO:0038156) |
| 0.2 | 0.5 | GO:0048058 | compound eye corneal lens development(GO:0048058) |
| 0.2 | 1.3 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.2 | 0.8 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.2 | 1.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 1.4 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 1.1 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 | 0.6 | GO:0003069 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 0.6 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.1 | 2.1 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.5 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.6 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 1.8 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 | 0.6 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.1 | 0.4 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.1 | 0.4 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.1 | 0.5 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.1 | 0.3 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.1 | 0.3 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.1 | 0.3 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.1 | 0.5 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.1 | 0.3 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.1 | 0.5 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 0.3 | GO:1901257 | negative regulation of interleukin-3 production(GO:0032712) negative regulation of granulocyte colony-stimulating factor production(GO:0071656) negative regulation of macrophage colony-stimulating factor production(GO:1901257) |
| 0.1 | 0.3 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.1 | 0.3 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 0.7 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.8 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.3 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.1 | 0.5 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 0.1 | 0.4 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 0.7 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.1 | 2.0 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 0.4 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.1 | 0.3 | GO:0035936 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.1 | 0.3 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.1 | 0.2 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 | 0.5 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.1 | 1.1 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.1 | 0.2 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.1 | 0.6 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 1.2 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.1 | 0.1 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.1 | 0.3 | GO:1902869 | regulation of amacrine cell differentiation(GO:1902869) |
| 0.1 | 0.3 | GO:0009213 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 0.1 | 0.2 | GO:0072276 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 0.6 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 0.2 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.1 | 0.3 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.1 | 0.9 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.1 | 0.4 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 0.5 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.2 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 0.1 | 0.4 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 0.7 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.2 | GO:1904692 | positive regulation of type B pancreatic cell proliferation(GO:1904692) |
| 0.1 | 0.3 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 0.2 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.6 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.2 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.1 | 0.7 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.1 | 0.4 | GO:0001712 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.1 | 0.4 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.1 | 0.2 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.2 | GO:0006408 | snRNA export from nucleus(GO:0006408) |
| 0.1 | 0.2 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 0.0 | 0.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.3 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.8 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.6 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.0 | 0.8 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) cell proliferation involved in heart valve development(GO:2000793) |
| 0.0 | 0.1 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.0 | 0.2 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.3 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.3 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.0 | GO:0003245 | cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.1 | GO:2001303 | regulation of engulfment of apoptotic cell(GO:1901074) lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.0 | 0.2 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.6 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 1.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.1 | GO:1902232 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) regulation of positive thymic T cell selection(GO:1902232) |
| 0.0 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.4 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.0 | 0.2 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.3 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.1 | GO:0090301 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.0 | 0.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.0 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.0 | 0.1 | GO:1905033 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.0 | 0.2 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.4 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.1 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.1 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.0 | 0.1 | GO:0035498 | carnosine metabolic process(GO:0035498) |
| 0.0 | 0.4 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) |
| 0.0 | 0.4 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.2 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) pyroptosis(GO:0070269) |
| 0.0 | 0.1 | GO:0018282 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.0 | 0.8 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 1.0 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.1 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.1 | GO:0072302 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.3 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.1 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 0.0 | 0.1 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 1.2 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.0 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.2 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.5 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.4 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 1.3 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.2 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.0 | 0.1 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 0.5 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) |
| 0.0 | 0.4 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.3 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.1 | GO:0061570 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.0 | 0.5 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.4 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.4 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.2 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.1 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.3 | GO:0032740 | positive regulation of interleukin-17 production(GO:0032740) |
| 0.0 | 0.1 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.1 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.7 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.2 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.3 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.5 | GO:0051450 | myoblast proliferation(GO:0051450) |
| 0.0 | 0.5 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.1 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.0 | 0.7 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.9 | GO:0035825 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) positive regulation of histone H3-K9 trimethylation(GO:1900114) |
| 0.0 | 1.9 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.3 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.0 | 0.3 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0006726 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.1 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.4 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.1 | GO:1903936 | cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.0 | GO:0036100 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.0 | 0.2 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 1.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0060029 | convergent extension involved in organogenesis(GO:0060029) |
| 0.0 | 0.0 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.0 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.4 | GO:0021756 | striatum development(GO:0021756) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.7 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.6 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.0 | GO:0071422 | succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) |
| 0.0 | 0.7 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.0 | 0.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.0 | 0.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.1 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.1 | GO:0032907 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) |
| 0.0 | 0.1 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.4 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.0 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.2 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.0 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.0 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.2 | GO:0035090 | telencephalon regionalization(GO:0021978) maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.2 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.3 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.1 | GO:0060026 | convergent extension(GO:0060026) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.2 | GO:0060742 | epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.0 | 0.2 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.0 | GO:1901053 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.3 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.3 | GO:0042825 | TAP complex(GO:0042825) |
| 0.2 | 1.4 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.2 | 1.7 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.4 | GO:0000229 | cytoplasmic chromosome(GO:0000229) |
| 0.1 | 1.0 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.1 | 0.4 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 0.3 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 0.3 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 1.5 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.5 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.4 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.7 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 2.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.3 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.1 | 0.6 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.3 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 0.5 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.2 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 0.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.2 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 0.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.3 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.2 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.6 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.3 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 3.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.4 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.1 | GO:0070701 | mucus layer(GO:0070701) |
| 0.0 | 0.2 | GO:0097169 | IPAF inflammasome complex(GO:0072557) AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.4 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.8 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.2 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.9 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0033648 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.4 | GO:0044615 | nuclear pore central transport channel(GO:0044613) nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.4 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.2 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 1.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.8 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.1 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 1.2 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.4 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 2.4 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 4.4 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.3 | 2.6 | GO:0023029 | MHC class Ib protein binding(GO:0023029) |
| 0.2 | 1.3 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.2 | 3.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 0.5 | GO:0004912 | interleukin-3 receptor activity(GO:0004912) interleukin-5 receptor activity(GO:0004914) |
| 0.2 | 1.0 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.2 | 0.5 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.2 | 0.6 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.2 | 1.7 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.2 | 0.5 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 1.0 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.5 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.1 | 0.5 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 0.6 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 0.3 | GO:0004040 | amidase activity(GO:0004040) |
| 0.1 | 0.3 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.1 | 0.3 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.1 | 0.9 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.5 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.5 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.1 | 0.2 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.1 | 0.4 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.1 | 0.4 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.1 | 0.5 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.2 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.1 | 0.3 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 0.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.6 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.3 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.1 | 0.2 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 0.1 | 0.8 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.2 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.1 | 1.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.4 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.4 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 1.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.2 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.1 | 0.2 | GO:0038047 | beta-endorphin receptor activity(GO:0004979) morphine receptor activity(GO:0038047) |
| 0.1 | 0.2 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.7 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 2.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.4 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.7 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.4 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.0 | 1.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.3 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.4 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.1 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.6 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.5 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 2.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 0.2 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 1.0 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.5 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.7 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.8 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 1.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.6 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.9 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.4 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 1.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.3 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.8 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.7 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 1.0 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 2.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.1 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.0 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.2 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0099589 | serotonin receptor activity(GO:0099589) |
| 0.0 | 0.1 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.4 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.2 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.0 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.0 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.9 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.9 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 2.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 2.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 2.2 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 1.5 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.9 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.2 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 3.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.5 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.3 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 3.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.6 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.9 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 2.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.8 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.9 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 1.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.1 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.9 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.1 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |