Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for OLIG3_NEUROD2_NEUROG2
Z-value: 0.73
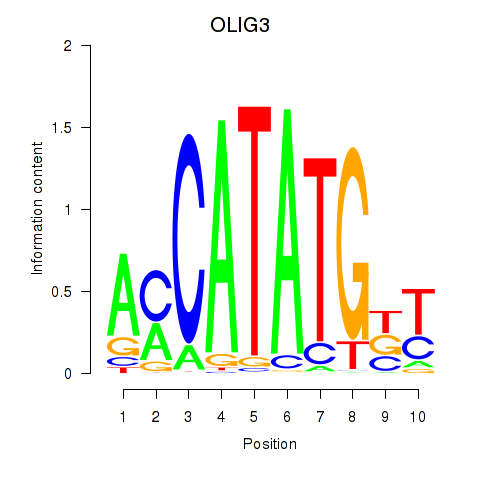
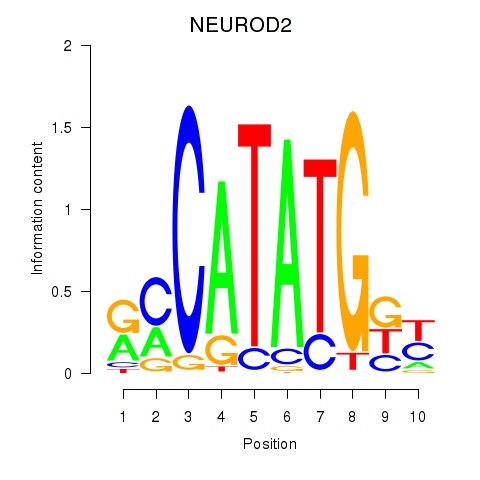
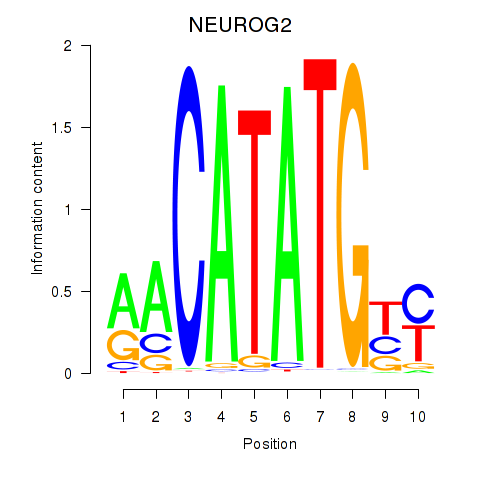
Transcription factors associated with OLIG3_NEUROD2_NEUROG2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OLIG3
|
ENSG00000177468.7 | OLIG3 |
|
NEUROD2
|
ENSG00000171532.5 | NEUROD2 |
|
NEUROG2
|
ENSG00000178403.4 | NEUROG2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| OLIG3 | hg38_v1_chr6_-_137494387_137494400 | -0.26 | 2.1e-01 | Click! |
| NEUROG2 | hg38_v1_chr4_-_112516176_112516181 | -0.19 | 3.6e-01 | Click! |
| NEUROD2 | hg38_v1_chr17_-_39607876_39607966 | -0.08 | 7.0e-01 | Click! |
Activity profile of OLIG3_NEUROD2_NEUROG2 motif
Sorted Z-values of OLIG3_NEUROD2_NEUROG2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of OLIG3_NEUROD2_NEUROG2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_92017637 | 5.00 |
ENST00000422361.6
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr3_-_18438767 | 4.93 |
ENST00000454909.6
|
SATB1
|
SATB homeobox 1 |
| chr19_-_11339573 | 4.02 |
ENST00000222120.8
|
RAB3D
|
RAB3D, member RAS oncogene family |
| chr1_+_99646025 | 4.00 |
ENST00000263174.9
ENST00000605497.5 ENST00000615664.1 |
PALMD
|
palmdelphin |
| chr5_-_111976925 | 2.38 |
ENST00000395634.7
|
NREP
|
neuronal regeneration related protein |
| chr7_-_11832190 | 1.92 |
ENST00000423059.9
ENST00000617773.1 |
THSD7A
|
thrombospondin type 1 domain containing 7A |
| chr1_-_153094521 | 1.75 |
ENST00000368750.8
|
SPRR2E
|
small proline rich protein 2E |
| chr3_-_197573323 | 1.71 |
ENST00000358186.6
ENST00000431056.5 |
BDH1
|
3-hydroxybutyrate dehydrogenase 1 |
| chr2_-_182242031 | 1.64 |
ENST00000358139.6
|
PDE1A
|
phosphodiesterase 1A |
| chr7_-_27165517 | 1.55 |
ENST00000396345.1
ENST00000343483.7 |
HOXA9
|
homeobox A9 |
| chr20_+_36214373 | 1.39 |
ENST00000432603.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr17_+_58692563 | 1.30 |
ENST00000461271.5
ENST00000583539.5 ENST00000337432.9 ENST00000421782.3 |
RAD51C
|
RAD51 paralog C |
| chr7_+_116862552 | 1.25 |
ENST00000361183.8
ENST00000639546.1 ENST00000490693.5 |
CAPZA2
|
capping actin protein of muscle Z-line subunit alpha 2 |
| chr10_+_5094405 | 1.21 |
ENST00000380554.5
|
AKR1C3
|
aldo-keto reductase family 1 member C3 |
| chr4_+_143433491 | 1.21 |
ENST00000512843.1
|
GAB1
|
GRB2 associated binding protein 1 |
| chrX_-_126552801 | 1.11 |
ENST00000371126.3
|
DCAF12L1
|
DDB1 and CUL4 associated factor 12 like 1 |
| chrX_+_118974608 | 1.08 |
ENST00000304778.11
ENST00000371628.8 |
LONRF3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr3_-_179266971 | 1.08 |
ENST00000349697.2
ENST00000497599.5 |
KCNMB3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr4_-_185812209 | 1.07 |
ENST00000393523.6
ENST00000393528.7 ENST00000449407.6 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr3_-_126357399 | 1.06 |
ENST00000296233.4
|
KLF15
|
Kruppel like factor 15 |
| chr4_-_158173004 | 1.03 |
ENST00000585682.6
|
GASK1B
|
golgi associated kinase 1B |
| chrX_-_63351308 | 1.00 |
ENST00000374884.3
|
SPIN4
|
spindlin family member 4 |
| chr2_-_187513641 | 0.98 |
ENST00000392365.5
ENST00000435414.5 |
TFPI
|
tissue factor pathway inhibitor |
| chr16_-_66549839 | 0.97 |
ENST00000527800.6
ENST00000677555.1 ENST00000563369.6 |
TK2
|
thymidine kinase 2 |
| chr8_-_38382146 | 0.97 |
ENST00000534155.1
ENST00000433384.6 ENST00000317025.13 ENST00000316985.7 |
NSD3
|
nuclear receptor binding SET domain protein 3 |
| chr9_+_76459152 | 0.95 |
ENST00000444201.6
ENST00000376730.5 |
GCNT1
|
glucosaminyl (N-acetyl) transferase 1 |
| chr2_-_113241779 | 0.93 |
ENST00000497038.6
|
PAX8
|
paired box 8 |
| chr2_-_182522703 | 0.91 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A |
| chr3_-_114759115 | 0.88 |
ENST00000471418.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr13_+_31945826 | 0.85 |
ENST00000647500.1
|
FRY
|
FRY microtubule binding protein |
| chr14_+_22207502 | 0.83 |
ENST00000390461.2
|
TRAV34
|
T cell receptor alpha variable 34 |
| chr1_+_244051275 | 0.80 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr3_-_149086488 | 0.80 |
ENST00000392912.6
ENST00000465259.5 ENST00000310053.10 ENST00000494055.5 |
HLTF
|
helicase like transcription factor |
| chr2_-_113241683 | 0.80 |
ENST00000468980.3
|
PAX8
|
paired box 8 |
| chr3_-_114758940 | 0.78 |
ENST00000464560.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr6_-_52994248 | 0.76 |
ENST00000457564.1
ENST00000370960.5 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr15_-_70763539 | 0.76 |
ENST00000322954.11
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr4_+_155666827 | 0.75 |
ENST00000511507.5
ENST00000506455.6 |
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr4_+_155666963 | 0.73 |
ENST00000455639.6
|
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr4_-_158173042 | 0.73 |
ENST00000592057.1
ENST00000393807.9 |
GASK1B
|
golgi associated kinase 1B |
| chr4_+_155666718 | 0.73 |
ENST00000621234.4
ENST00000511108.5 |
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr7_-_113118544 | 0.73 |
ENST00000397764.8
ENST00000441359.1 |
SMIM30
|
small integral membrane protein 30 |
| chr3_-_108953762 | 0.73 |
ENST00000393963.7
ENST00000471108.1 |
GUCA1C
|
guanylate cyclase activator 1C |
| chr15_-_55408018 | 0.72 |
ENST00000569205.5
|
CCPG1
|
cell cycle progression 1 |
| chr15_+_65550819 | 0.72 |
ENST00000569894.5
|
HACD3
|
3-hydroxyacyl-CoA dehydratase 3 |
| chr18_+_79863668 | 0.71 |
ENST00000316249.3
|
KCNG2
|
potassium voltage-gated channel modifier subfamily G member 2 |
| chr4_+_113292838 | 0.70 |
ENST00000672411.1
ENST00000673231.1 |
ANK2
|
ankyrin 2 |
| chr5_+_149581368 | 0.69 |
ENST00000333677.7
|
ARHGEF37
|
Rho guanine nucleotide exchange factor 37 |
| chr6_+_2999885 | 0.68 |
ENST00000397717.7
ENST00000380455.11 |
NQO2
|
N-ribosyldihydronicotinamide:quinone reductase 2 |
| chr12_+_64610458 | 0.68 |
ENST00000542104.6
|
RASSF3
|
Ras association domain family member 3 |
| chr2_+_71453722 | 0.67 |
ENST00000409582.7
ENST00000409762.5 ENST00000413539.6 ENST00000429174.6 |
DYSF
|
dysferlin |
| chr12_+_51048318 | 0.65 |
ENST00000550929.5
ENST00000550442.5 ENST00000549340.5 ENST00000548209.5 ENST00000548251.5 ENST00000550814.5 ENST00000547660.5 ENST00000262055.9 ENST00000548401.1 ENST00000418425.6 ENST00000547008.5 ENST00000552739.5 |
LETMD1
|
LETM1 domain containing 1 |
| chr4_-_149815826 | 0.65 |
ENST00000636793.2
ENST00000636414.1 |
IQCM
|
IQ motif containing M |
| chr2_+_190927649 | 0.65 |
ENST00000409428.5
ENST00000409215.5 |
GLS
|
glutaminase |
| chr1_+_248508073 | 0.64 |
ENST00000641804.1
|
OR2G6
|
olfactory receptor family 2 subfamily G member 6 |
| chr4_+_113292925 | 0.64 |
ENST00000673353.1
ENST00000505342.6 ENST00000672915.1 ENST00000509550.5 |
ANK2
|
ankyrin 2 |
| chr6_+_2999961 | 0.64 |
ENST00000338130.7
|
NQO2
|
N-ribosyldihydronicotinamide:quinone reductase 2 |
| chr3_-_108953870 | 0.64 |
ENST00000261047.8
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr2_-_105396943 | 0.63 |
ENST00000409807.5
|
FHL2
|
four and a half LIM domains 2 |
| chr7_-_38363476 | 0.63 |
ENST00000426402.2
|
TRGV2
|
T cell receptor gamma variable 2 |
| chr6_-_75363003 | 0.63 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr15_-_65211463 | 0.62 |
ENST00000261883.6
|
CILP
|
cartilage intermediate layer protein |
| chr6_-_30744537 | 0.62 |
ENST00000259874.6
ENST00000376377.2 |
IER3
|
immediate early response 3 |
| chr5_-_115296610 | 0.61 |
ENST00000379611.10
ENST00000506442.5 |
CCDC112
|
coiled-coil domain containing 112 |
| chr6_-_2744126 | 0.61 |
ENST00000647417.1
|
MYLK4
|
myosin light chain kinase family member 4 |
| chr17_+_7834200 | 0.60 |
ENST00000448097.7
|
KDM6B
|
lysine demethylase 6B |
| chr6_+_30888672 | 0.60 |
ENST00000446312.5
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr3_+_37975773 | 0.59 |
ENST00000436654.1
|
CTDSPL
|
CTD small phosphatase like |
| chr4_-_88823306 | 0.58 |
ENST00000395002.6
|
FAM13A
|
family with sequence similarity 13 member A |
| chr3_-_43105939 | 0.58 |
ENST00000441964.1
|
POMGNT2
|
protein O-linked mannose N-acetylglucosaminyltransferase 2 (beta 1,4-) |
| chr5_+_175872741 | 0.57 |
ENST00000502265.5
|
CPLX2
|
complexin 2 |
| chr15_+_84600986 | 0.57 |
ENST00000540936.1
ENST00000448803.6 ENST00000546275.1 ENST00000546148.6 ENST00000442073.3 ENST00000334141.7 ENST00000358472.3 ENST00000502939.2 ENST00000379358.7 ENST00000327179.6 |
ZSCAN2
|
zinc finger and SCAN domain containing 2 |
| chr19_+_11925062 | 0.57 |
ENST00000622593.4
ENST00000590798.1 |
ZNF700
ENSG00000267179.1
|
zinc finger protein 700 novel protein |
| chr9_+_2158239 | 0.56 |
ENST00000635133.1
ENST00000634931.1 ENST00000423555.6 ENST00000382185.6 ENST00000302401.8 ENST00000382183.6 ENST00000417599.6 ENST00000382186.6 ENST00000635530.1 ENST00000635388.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr22_+_46674593 | 0.56 |
ENST00000408031.1
|
GRAMD4
|
GRAM domain containing 4 |
| chr2_+_233059838 | 0.55 |
ENST00000359570.9
|
INPP5D
|
inositol polyphosphate-5-phosphatase D |
| chr19_+_11925098 | 0.55 |
ENST00000254321.10
ENST00000591944.1 |
ZNF700
ENSG00000267179.1
|
zinc finger protein 700 novel protein |
| chr1_+_148748774 | 0.54 |
ENST00000322209.5
|
NUDT4B
|
nudix hydrolase 4B |
| chr9_-_109013483 | 0.53 |
ENST00000325551.9
ENST00000374593.4 ENST00000374595.8 |
CTNNAL1
|
catenin alpha like 1 |
| chr6_+_2999984 | 0.52 |
ENST00000380441.5
ENST00000380454.8 |
NQO2
|
N-ribosyldihydronicotinamide:quinone reductase 2 |
| chr8_-_126558461 | 0.52 |
ENST00000304916.4
|
LRATD2
|
LRAT domain containing 2 |
| chr9_-_72953047 | 0.52 |
ENST00000297785.8
ENST00000376939.5 |
ALDH1A1
|
aldehyde dehydrogenase 1 family member A1 |
| chr12_-_102478539 | 0.51 |
ENST00000424202.6
|
IGF1
|
insulin like growth factor 1 |
| chr3_-_149377637 | 0.51 |
ENST00000305366.8
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr4_+_70734419 | 0.51 |
ENST00000502653.5
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr22_+_22899481 | 0.50 |
ENST00000390322.2
|
IGLJ2
|
immunoglobulin lambda joining 2 |
| chr1_-_21050952 | 0.49 |
ENST00000264211.12
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma 3 |
| chr17_-_49848017 | 0.49 |
ENST00000326219.5
ENST00000334568.8 ENST00000352793.6 ENST00000398154.5 ENST00000436235.5 |
TAC4
|
tachykinin precursor 4 |
| chr8_-_17895487 | 0.48 |
ENST00000427924.5
ENST00000381841.4 |
FGL1
|
fibrinogen like 1 |
| chr17_-_5123102 | 0.48 |
ENST00000250076.7
|
ZNF232
|
zinc finger protein 232 |
| chr15_+_57219411 | 0.47 |
ENST00000543579.5
ENST00000537840.5 ENST00000343827.7 |
TCF12
|
transcription factor 12 |
| chr15_-_63156774 | 0.47 |
ENST00000462430.5
|
RPS27L
|
ribosomal protein S27 like |
| chr11_-_134253248 | 0.46 |
ENST00000392595.6
ENST00000352327.5 ENST00000341541.8 ENST00000392594.7 |
THYN1
|
thymocyte nuclear protein 1 |
| chr11_+_134253531 | 0.46 |
ENST00000374752.6
ENST00000281182.9 |
ACAD8
|
acyl-CoA dehydrogenase family member 8 |
| chr17_+_68515399 | 0.46 |
ENST00000588188.6
|
PRKAR1A
|
protein kinase cAMP-dependent type I regulatory subunit alpha |
| chr14_-_74612226 | 0.45 |
ENST00000261978.9
|
LTBP2
|
latent transforming growth factor beta binding protein 2 |
| chr2_+_178320228 | 0.45 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein like 6 |
| chr3_-_50350710 | 0.45 |
ENST00000232501.8
|
NPRL2
|
NPR2 like, GATOR1 complex subunit |
| chr17_+_65137408 | 0.44 |
ENST00000443584.7
ENST00000449996.7 |
RGS9
|
regulator of G protein signaling 9 |
| chr2_+_71453538 | 0.44 |
ENST00000258104.8
|
DYSF
|
dysferlin |
| chr11_+_17295322 | 0.44 |
ENST00000458064.6
ENST00000622082.4 |
NUCB2
|
nucleobindin 2 |
| chr19_+_49691103 | 0.43 |
ENST00000323446.9
ENST00000392518.8 ENST00000598293.6 ENST00000598396.5 ENST00000405931.6 ENST00000602019.5 |
CPT1C
|
carnitine palmitoyltransferase 1C |
| chr6_-_118651522 | 0.43 |
ENST00000368491.8
|
CEP85L
|
centrosomal protein 85 like |
| chr15_+_40807055 | 0.42 |
ENST00000570108.5
ENST00000564258.5 ENST00000355341.8 ENST00000336455.9 |
ZFYVE19
|
zinc finger FYVE-type containing 19 |
| chrX_+_47585212 | 0.42 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr6_+_292050 | 0.42 |
ENST00000344450.9
|
DUSP22
|
dual specificity phosphatase 22 |
| chr9_-_124941054 | 0.41 |
ENST00000373555.9
|
GOLGA1
|
golgin A1 |
| chr9_+_5450503 | 0.41 |
ENST00000381573.8
ENST00000381577.4 |
CD274
|
CD274 molecule |
| chr15_+_75647877 | 0.41 |
ENST00000308527.6
|
SNX33
|
sorting nexin 33 |
| chr22_-_28306645 | 0.40 |
ENST00000612946.4
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr17_+_75633420 | 0.40 |
ENST00000375215.3
|
SMIM5
|
small integral membrane protein 5 |
| chr11_+_32091065 | 0.40 |
ENST00000054950.4
|
RCN1
|
reticulocalbin 1 |
| chr8_+_22057857 | 0.40 |
ENST00000517305.4
ENST00000265800.9 ENST00000517418.5 |
DMTN
|
dematin actin binding protein |
| chr11_+_7485492 | 0.39 |
ENST00000534244.1
ENST00000329293.4 |
OLFML1
|
olfactomedin like 1 |
| chr19_+_24087147 | 0.39 |
ENST00000611359.3
ENST00000616028.2 |
ZNF254
|
zinc finger protein 254 |
| chr6_-_118710065 | 0.39 |
ENST00000392500.7
ENST00000368488.9 ENST00000434604.5 |
CEP85L
|
centrosomal protein 85 like |
| chr7_+_148590760 | 0.39 |
ENST00000307003.3
|
C7orf33
|
chromosome 7 open reading frame 33 |
| chr14_-_106025628 | 0.38 |
ENST00000631943.1
|
IGHV7-4-1
|
immunoglobulin heavy variable 7-4-1 |
| chr11_-_62591500 | 0.38 |
ENST00000476907.6
ENST00000278279.7 |
TUT1
|
terminal uridylyl transferase 1, U6 snRNA-specific |
| chrX_-_10576901 | 0.38 |
ENST00000380779.5
|
MID1
|
midline 1 |
| chr2_+_177392734 | 0.38 |
ENST00000680770.1
ENST00000637633.2 ENST00000679459.1 ENST00000409888.1 ENST00000264167.11 ENST00000642466.2 |
AGPS
|
alkylglycerone phosphate synthase |
| chr4_-_73223082 | 0.38 |
ENST00000509867.6
|
ANKRD17
|
ankyrin repeat domain 17 |
| chr11_+_121590388 | 0.38 |
ENST00000527934.1
|
SORL1
|
sortilin related receptor 1 |
| chr16_+_53099100 | 0.38 |
ENST00000565832.5
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr15_+_49155750 | 0.37 |
ENST00000327171.7
ENST00000560654.5 |
GALK2
|
galactokinase 2 |
| chr4_-_24912954 | 0.37 |
ENST00000502801.1
ENST00000635206.2 |
CCDC149
|
coiled-coil domain containing 149 |
| chr8_+_21966215 | 0.37 |
ENST00000433566.8
|
XPO7
|
exportin 7 |
| chr3_-_186806445 | 0.37 |
ENST00000418288.5
ENST00000296273.7 |
RFC4
|
replication factor C subunit 4 |
| chr14_-_23435652 | 0.37 |
ENST00000355349.4
|
MYH7
|
myosin heavy chain 7 |
| chr18_+_13218769 | 0.37 |
ENST00000677055.1
ENST00000399848.7 |
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr12_-_54295748 | 0.37 |
ENST00000540264.2
ENST00000312156.8 |
NFE2
|
nuclear factor, erythroid 2 |
| chr8_-_134510182 | 0.36 |
ENST00000521673.5
|
ZFAT
|
zinc finger and AT-hook domain containing |
| chr2_+_180981108 | 0.36 |
ENST00000602710.5
|
UBE2E3
|
ubiquitin conjugating enzyme E2 E3 |
| chr2_+_97669739 | 0.36 |
ENST00000599501.6
ENST00000627399.3 ENST00000627284.2 ENST00000599435.5 ENST00000597654.5 ENST00000598737.5 |
C2orf92
|
chromosome 2 open reading frame 92 |
| chr6_-_28336375 | 0.36 |
ENST00000611469.4
ENST00000435857.5 |
ZSCAN31
|
zinc finger and SCAN domain containing 31 |
| chr6_-_31680377 | 0.36 |
ENST00000383237.4
|
LY6G5C
|
lymphocyte antigen 6 family member G5C |
| chr11_-_62591554 | 0.36 |
ENST00000494385.1
ENST00000308436.11 |
TUT1
|
terminal uridylyl transferase 1, U6 snRNA-specific |
| chr5_+_42548043 | 0.35 |
ENST00000618088.4
ENST00000612382.4 |
GHR
|
growth hormone receptor |
| chr3_-_43106057 | 0.35 |
ENST00000344697.3
|
POMGNT2
|
protein O-linked mannose N-acetylglucosaminyltransferase 2 (beta 1,4-) |
| chrX_+_108091665 | 0.35 |
ENST00000345734.7
|
ATG4A
|
autophagy related 4A cysteine peptidase |
| chr5_-_76623391 | 0.35 |
ENST00000296641.5
ENST00000504899.1 |
F2RL2
|
coagulation factor II thrombin receptor like 2 |
| chr9_+_2110354 | 0.35 |
ENST00000634772.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr12_-_52703522 | 0.35 |
ENST00000341809.8
|
KRT77
|
keratin 77 |
| chr20_-_5119945 | 0.34 |
ENST00000379143.10
|
PCNA
|
proliferating cell nuclear antigen |
| chr4_+_25914275 | 0.34 |
ENST00000514384.1
|
SMIM20
|
small integral membrane protein 20 |
| chr3_+_156674579 | 0.34 |
ENST00000295924.12
|
TIPARP
|
TCDD inducible poly(ADP-ribose) polymerase |
| chr1_-_16018005 | 0.33 |
ENST00000406363.2
ENST00000411503.5 ENST00000311890.14 ENST00000487046.1 |
HSPB7
|
heat shock protein family B (small) member 7 |
| chr11_+_118304881 | 0.33 |
ENST00000528600.1
|
CD3E
|
CD3e molecule |
| chr1_-_186375671 | 0.33 |
ENST00000451586.1
|
TPR
|
translocated promoter region, nuclear basket protein |
| chr2_+_33134620 | 0.32 |
ENST00000402934.5
ENST00000404525.5 ENST00000407925.5 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chrX_+_106611930 | 0.32 |
ENST00000372544.6
ENST00000372548.9 |
RADX
|
RPA1 related single stranded DNA binding protein, X-linked |
| chr11_-_60855943 | 0.32 |
ENST00000332539.5
|
PTGDR2
|
prostaglandin D2 receptor 2 |
| chr6_-_28336123 | 0.32 |
ENST00000439158.5
ENST00000446474.5 ENST00000414431.1 ENST00000344279.11 ENST00000453745.5 |
ZSCAN31
|
zinc finger and SCAN domain containing 31 |
| chr4_+_2812259 | 0.32 |
ENST00000502260.5
ENST00000435136.8 |
SH3BP2
|
SH3 domain binding protein 2 |
| chr11_+_118304721 | 0.32 |
ENST00000361763.9
|
CD3E
|
CD3e molecule |
| chr2_-_182522556 | 0.32 |
ENST00000435564.5
|
PDE1A
|
phosphodiesterase 1A |
| chr5_+_170861990 | 0.31 |
ENST00000523189.6
|
RANBP17
|
RAN binding protein 17 |
| chr2_+_33134579 | 0.31 |
ENST00000418533.6
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr17_+_47253817 | 0.31 |
ENST00000559488.7
ENST00000571680.1 |
ITGB3
|
integrin subunit beta 3 |
| chr3_+_119579577 | 0.31 |
ENST00000478927.5
|
ADPRH
|
ADP-ribosylarginine hydrolase |
| chr5_+_141192330 | 0.31 |
ENST00000239446.6
|
PCDHB10
|
protocadherin beta 10 |
| chr3_-_33659441 | 0.31 |
ENST00000650653.1
ENST00000480013.6 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr20_-_35529618 | 0.30 |
ENST00000246199.5
ENST00000424444.1 ENST00000374345.8 ENST00000444723.3 |
C20orf173
|
chromosome 20 open reading frame 173 |
| chr3_-_98522514 | 0.30 |
ENST00000503004.5
ENST00000506575.1 ENST00000513452.5 ENST00000515620.5 |
CLDND1
|
claudin domain containing 1 |
| chr5_+_108924585 | 0.30 |
ENST00000618353.1
|
FER
|
FER tyrosine kinase |
| chr10_-_27100463 | 0.30 |
ENST00000436985.7
ENST00000376087.5 |
ANKRD26
|
ankyrin repeat domain 26 |
| chr5_+_157180816 | 0.29 |
ENST00000422843.8
|
ITK
|
IL2 inducible T cell kinase |
| chr9_-_13175824 | 0.29 |
ENST00000545857.5
|
MPDZ
|
multiple PDZ domain crumbs cell polarity complex component |
| chr16_+_11965193 | 0.29 |
ENST00000053243.6
ENST00000396495.3 |
TNFRSF17
|
TNF receptor superfamily member 17 |
| chr4_+_118034480 | 0.29 |
ENST00000296499.6
|
NDST3
|
N-deacetylase and N-sulfotransferase 3 |
| chr17_+_19648915 | 0.28 |
ENST00000672567.1
ENST00000672709.1 |
ALDH3A2
|
aldehyde dehydrogenase 3 family member A2 |
| chr10_-_112183698 | 0.28 |
ENST00000369425.5
ENST00000348367.9 |
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr1_-_77219399 | 0.28 |
ENST00000359130.1
ENST00000370812.8 ENST00000445065.5 |
PIGK
|
phosphatidylinositol glycan anchor biosynthesis class K |
| chr19_-_51723968 | 0.28 |
ENST00000222115.5
ENST00000540069.7 |
HAS1
|
hyaluronan synthase 1 |
| chr16_+_53054973 | 0.28 |
ENST00000447540.6
ENST00000615216.4 ENST00000566029.5 |
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr17_-_67996428 | 0.28 |
ENST00000580729.3
|
C17orf58
|
chromosome 17 open reading frame 58 |
| chr3_-_33659097 | 0.28 |
ENST00000461133.8
ENST00000496954.2 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr6_+_28124596 | 0.27 |
ENST00000340487.5
|
ZSCAN16
|
zinc finger and SCAN domain containing 16 |
| chr6_+_72366730 | 0.27 |
ENST00000414192.2
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr2_-_183038405 | 0.27 |
ENST00000361354.9
|
NCKAP1
|
NCK associated protein 1 |
| chr17_-_60392333 | 0.27 |
ENST00000590133.5
|
USP32
|
ubiquitin specific peptidase 32 |
| chr5_+_149357999 | 0.27 |
ENST00000274569.9
|
PCYOX1L
|
prenylcysteine oxidase 1 like |
| chr10_+_23095556 | 0.27 |
ENST00000376510.8
|
MSRB2
|
methionine sulfoxide reductase B2 |
| chr1_+_159302321 | 0.27 |
ENST00000368114.1
|
FCER1A
|
Fc fragment of IgE receptor Ia |
| chr17_-_60392113 | 0.27 |
ENST00000300896.9
ENST00000589335.5 |
USP32
|
ubiquitin specific peptidase 32 |
| chr19_-_20039492 | 0.27 |
ENST00000397165.7
|
ZNF682
|
zinc finger protein 682 |
| chr2_+_74554692 | 0.27 |
ENST00000233668.10
ENST00000340004.6 |
DOK1
|
docking protein 1 |
| chr1_+_74198230 | 0.27 |
ENST00000557284.7
ENST00000370899.7 ENST00000370895.5 ENST00000534632.5 ENST00000370893.1 ENST00000467578.7 ENST00000370894.9 ENST00000482102.2 ENST00000370898.9 ENST00000534056.5 |
FPGT-TNNI3K
FPGT
|
FPGT-TNNI3K readthrough fucose-1-phosphate guanylyltransferase |
| chr7_-_156892987 | 0.26 |
ENST00000415428.5
|
LMBR1
|
limb development membrane protein 1 |
| chr3_+_46883337 | 0.26 |
ENST00000313049.9
|
PTH1R
|
parathyroid hormone 1 receptor |
| chr20_-_31406207 | 0.26 |
ENST00000376314.3
|
DEFB121
|
defensin beta 121 |
| chrX_+_136196750 | 0.26 |
ENST00000539015.5
|
FHL1
|
four and a half LIM domains 1 |
| chr10_-_48251757 | 0.25 |
ENST00000305531.3
|
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr2_+_105337515 | 0.25 |
ENST00000410049.1
ENST00000258457.7 |
C2orf49
|
chromosome 2 open reading frame 49 |
| chr14_+_24429932 | 0.25 |
ENST00000556842.5
ENST00000553935.6 |
KHNYN
|
KH and NYN domain containing |
| chr9_+_18474165 | 0.25 |
ENST00000380566.8
|
ADAMTSL1
|
ADAMTS like 1 |
| chr2_+_209580024 | 0.25 |
ENST00000392194.5
|
MAP2
|
microtubule associated protein 2 |
| chr12_-_10884244 | 0.25 |
ENST00000543626.4
|
PRH1
|
proline rich protein HaeIII subfamily 1 |
| chr11_+_118359572 | 0.25 |
ENST00000252108.8
ENST00000431736.6 |
UBE4A
|
ubiquitination factor E4A |
| chr18_-_50287816 | 0.24 |
ENST00000589548.6
ENST00000673786.1 |
CXXC1
|
CXXC finger protein 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.0 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.4 | 1.7 | GO:2000612 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.4 | 1.2 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.3 | 5.0 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.3 | 1.7 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.2 | 0.8 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.2 | 1.4 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.2 | 1.0 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.2 | 0.6 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.2 | 0.6 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.6 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 1.3 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 2.9 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.1 | 0.4 | GO:1902997 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 | 0.5 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.1 | 1.0 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.3 | GO:0031445 | regulation of heterochromatin assembly(GO:0031445) positive regulation of heterochromatin assembly(GO:0031453) |
| 0.1 | 0.6 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.3 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.1 | 0.5 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.1 | 0.6 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 0.9 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.7 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.1 | 0.3 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 0.3 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.1 | 0.6 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.3 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.1 | 0.3 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.1 | 0.3 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 1.4 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 0.6 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.2 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.1 | 0.9 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.1 | 0.2 | GO:0019049 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.5 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.4 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.4 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.3 | GO:0038109 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) diapedesis(GO:0050904) |
| 0.1 | 0.5 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.1 | 0.2 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.1 | 0.2 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.1 | 0.3 | GO:1903173 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.1 | 0.3 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.4 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.1 | 1.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.4 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.7 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.2 | GO:0045553 | TRAIL biosynthetic process(GO:0045553) regulation of TRAIL biosynthetic process(GO:0045554) positive regulation of TRAIL biosynthetic process(GO:0045556) |
| 0.1 | 0.2 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.1 | 0.6 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.3 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.1 | 0.2 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.1 | 1.7 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.1 | 0.2 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.3 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.1 | 0.4 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 0.9 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 1.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.4 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.2 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 0.3 | GO:0032902 | nerve growth factor production(GO:0032902) |
| 0.1 | 0.2 | GO:0003064 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.4 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 1.0 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.4 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.4 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.2 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 1.1 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.6 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.1 | GO:0036111 | very long-chain fatty-acyl-CoA metabolic process(GO:0036111) |
| 0.0 | 0.5 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.3 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 1.2 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 1.5 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.6 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.2 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.0 | 0.1 | GO:0050993 | dimethylallyl diphosphate biosynthetic process(GO:0050992) dimethylallyl diphosphate metabolic process(GO:0050993) |
| 0.0 | 0.4 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.6 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.2 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.0 | 0.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.2 | GO:0046103 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.0 | 2.0 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 1.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:1901377 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 2.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 1.1 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) |
| 0.0 | 0.1 | GO:0002881 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.4 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0006581 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.4 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.0 | 0.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.3 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.8 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 1.4 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:1903939 | regulation of TORC2 signaling(GO:1903939) |
| 0.0 | 0.2 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 0.4 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.2 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.3 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.2 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:1903249 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) negative regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901205) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.4 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.1 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.1 | GO:0010607 | negative regulation of cytoplasmic mRNA processing body assembly(GO:0010607) |
| 0.0 | 0.3 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.2 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.0 | 0.3 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.4 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.1 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.2 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.2 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.1 | GO:0072757 | cellular response to camptothecin(GO:0072757) |
| 0.0 | 0.3 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.1 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 | 0.5 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 1.0 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.4 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.0 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.4 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.1 | GO:0015783 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.1 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.0 | 0.1 | GO:0046398 | UDP-glucuronate biosynthetic process(GO:0006065) UDP-glucuronate metabolic process(GO:0046398) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.0 | 0.2 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.0 | 0.3 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.1 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.0 | 0.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.0 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.3 | GO:0060732 | regulation of inositol phosphate biosynthetic process(GO:0010919) positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.1 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 0.5 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.3 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.2 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.0 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.0 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.3 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.2 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.4 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.1 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 0.3 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.0 | 0.1 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.2 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.7 | GO:0006301 | postreplication repair(GO:0006301) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 3.6 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 0.4 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 0.3 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 4.9 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 0.3 | GO:0036027 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.1 | 0.6 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.1 | 1.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.7 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.5 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 0.6 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 1.8 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 0.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.6 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.7 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 1.7 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.0 | 0.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 1.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.8 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.4 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.5 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 3.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.1 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.3 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 1.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.3 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 3.7 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.9 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.2 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.1 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 1.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 1.4 | GO:0043204 | perikaryon(GO:0043204) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.5 | 2.9 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.5 | 1.8 | GO:0001512 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.4 | 1.2 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.3 | 0.9 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.3 | 0.9 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.2 | 1.7 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.2 | 0.3 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.2 | 1.4 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 1.8 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.0 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.1 | 1.4 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.4 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.1 | 3.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.5 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.6 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 0.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.3 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.1 | 0.7 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.7 | GO:0102345 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 0.6 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.3 | GO:0046577 | long-chain-alcohol oxidase activity(GO:0046577) |
| 0.1 | 0.6 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.6 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.5 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 0.6 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.4 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.1 | 0.2 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.6 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.2 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 0.3 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 0.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.8 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.2 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 0.3 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.1 | 0.3 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 0.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.2 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.1 | 0.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.6 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.2 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.1 | 0.2 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.4 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.3 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.4 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.4 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.2 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.1 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 1.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.0 | 0.1 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.0 | 0.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.1 | GO:0001626 | nociceptin receptor activity(GO:0001626) |
| 0.0 | 0.5 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.3 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.3 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0098519 | phosphoserine phosphatase activity(GO:0004647) nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.7 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.1 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.2 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.5 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 1.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.9 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0034617 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.0 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.0 | 0.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.2 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.1 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 1.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0001132 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.0 | 0.1 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.3 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 1.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 5.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 4.7 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.0 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.9 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.1 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.0 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.1 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.1 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 5.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 2.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 2.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.8 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.5 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.5 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.9 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 5.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.5 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.7 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 1.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 1.0 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.9 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 1.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 1.5 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.8 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.9 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.1 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.4 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.1 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |