Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
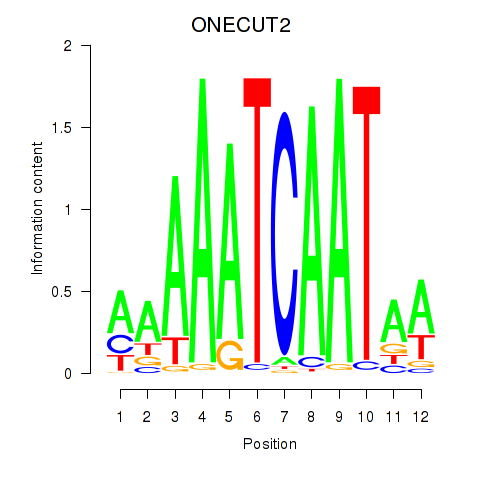
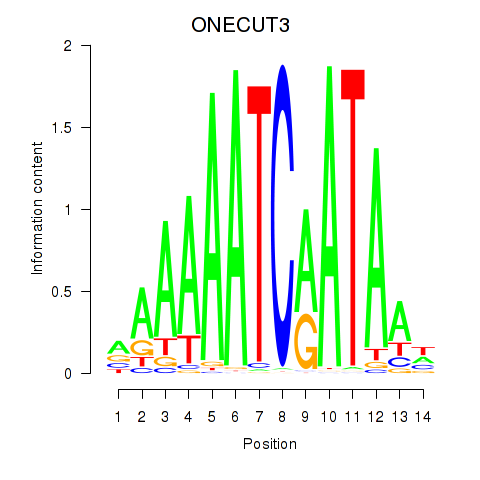
Results for ONECUT2_ONECUT3
Z-value: 0.47
Transcription factors associated with ONECUT2_ONECUT3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ONECUT2
|
ENSG00000119547.6 | ONECUT2 |
|
ONECUT3
|
ENSG00000205922.5 | ONECUT3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ONECUT2 | hg38_v1_chr18_+_57435366_57435391 | 0.43 | 3.4e-02 | Click! |
| ONECUT3 | hg38_v1_chr19_+_1753499_1753521 | -0.01 | 9.5e-01 | Click! |
Activity profile of ONECUT2_ONECUT3 motif
Sorted Z-values of ONECUT2_ONECUT3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ONECUT2_ONECUT3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_207165923 | 1.38 |
ENST00000309446.11
|
KLF7
|
Kruppel like factor 7 |
| chr5_+_141421064 | 0.71 |
ENST00000518882.2
|
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr4_-_69961007 | 0.69 |
ENST00000353151.3
|
CSN2
|
casein beta |
| chr11_-_102780620 | 0.66 |
ENST00000279441.9
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 |
| chr6_-_154430495 | 0.40 |
ENST00000424998.3
|
CNKSR3
|
CNKSR family member 3 |
| chr8_+_31639755 | 0.39 |
ENST00000520407.5
|
NRG1
|
neuregulin 1 |
| chr7_-_93148345 | 0.37 |
ENST00000437805.5
ENST00000446959.5 ENST00000439952.5 ENST00000414791.5 ENST00000446033.1 ENST00000411955.5 ENST00000318238.9 |
SAMD9L
|
sterile alpha motif domain containing 9 like |
| chr1_-_686673 | 0.37 |
ENST00000332831.4
|
OR4F16
|
olfactory receptor family 4 subfamily F member 16 |
| chr11_+_63369779 | 0.37 |
ENST00000279178.4
|
SLC22A9
|
solute carrier family 22 member 9 |
| chr5_+_181367268 | 0.34 |
ENST00000456475.1
|
OR4F3
|
olfactory receptor family 4 subfamily F member 3 |
| chr16_+_56191476 | 0.34 |
ENST00000262493.12
|
GNAO1
|
G protein subunit alpha o1 |
| chr14_+_36657560 | 0.33 |
ENST00000402703.6
|
PAX9
|
paired box 9 |
| chr6_+_29100609 | 0.32 |
ENST00000377171.3
|
OR2J1
|
olfactory receptor family 2 subfamily J member 1 |
| chr8_-_167024 | 0.32 |
ENST00000320901.4
|
OR4F21
|
olfactory receptor family 4 subfamily F member 21 |
| chr16_+_25216943 | 0.31 |
ENST00000219660.6
|
AQP8
|
aquaporin 8 |
| chr1_-_110607816 | 0.30 |
ENST00000485317.6
|
KCNA2
|
potassium voltage-gated channel subfamily A member 2 |
| chr1_-_110607970 | 0.30 |
ENST00000638532.1
|
KCNA2
|
potassium voltage-gated channel subfamily A member 2 |
| chr14_-_23578756 | 0.30 |
ENST00000397118.7
ENST00000356300.9 |
JPH4
|
junctophilin 4 |
| chr16_-_66918839 | 0.30 |
ENST00000565235.2
ENST00000568632.5 ENST00000565796.5 |
CDH16
|
cadherin 16 |
| chr16_-_66918876 | 0.30 |
ENST00000570262.5
ENST00000299752.9 ENST00000394055.7 |
CDH16
|
cadherin 16 |
| chr1_-_243163310 | 0.28 |
ENST00000492145.1
ENST00000490813.5 ENST00000464936.5 |
CEP170
|
centrosomal protein 170 |
| chr11_-_58844484 | 0.27 |
ENST00000532258.1
|
GLYATL2
|
glycine-N-acyltransferase like 2 |
| chr5_-_160852200 | 0.27 |
ENST00000327245.10
|
ATP10B
|
ATPase phospholipid transporting 10B (putative) |
| chr12_-_62192762 | 0.27 |
ENST00000416284.8
|
TAFA2
|
TAFA chemokine like family member 2 |
| chr14_+_22543179 | 0.27 |
ENST00000390534.1
|
TRAJ3
|
T cell receptor alpha joining 3 |
| chr8_+_32647080 | 0.27 |
ENST00000520502.7
ENST00000523041.2 ENST00000650819.1 |
NRG1
|
neuregulin 1 |
| chr2_-_24328113 | 0.25 |
ENST00000622089.4
|
ITSN2
|
intersectin 2 |
| chr5_-_116574802 | 0.23 |
ENST00000343348.11
|
SEMA6A
|
semaphorin 6A |
| chr19_+_9087061 | 0.23 |
ENST00000641627.1
|
OR1M1
|
olfactory receptor family 1 subfamily M member 1 |
| chr3_-_71581540 | 0.23 |
ENST00000650068.1
|
FOXP1
|
forkhead box P1 |
| chr3_+_63443076 | 0.23 |
ENST00000295894.9
|
SYNPR
|
synaptoporin |
| chr17_+_7558296 | 0.22 |
ENST00000438470.5
ENST00000436057.5 |
TNFSF13
|
TNF superfamily member 13 |
| chr8_+_53851786 | 0.22 |
ENST00000297313.8
ENST00000344277.10 |
RGS20
|
regulator of G protein signaling 20 |
| chr17_+_32991844 | 0.21 |
ENST00000269053.8
|
SPACA3
|
sperm acrosome associated 3 |
| chr7_+_43112593 | 0.20 |
ENST00000453890.5
ENST00000395891.7 |
HECW1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr11_-_58844695 | 0.20 |
ENST00000287275.6
|
GLYATL2
|
glycine-N-acyltransferase like 2 |
| chr5_+_141421020 | 0.20 |
ENST00000622044.1
ENST00000398587.7 |
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr2_+_54558348 | 0.20 |
ENST00000333896.5
|
SPTBN1
|
spectrin beta, non-erythrocytic 1 |
| chr19_+_3880647 | 0.19 |
ENST00000450849.7
|
ATCAY
|
ATCAY kinesin light chain interacting caytaxin |
| chr6_+_113857333 | 0.18 |
ENST00000612661.2
|
MARCKS
|
myristoylated alanine rich protein kinase C substrate |
| chr6_+_34236865 | 0.18 |
ENST00000674029.1
ENST00000447654.5 ENST00000347617.10 ENST00000401473.7 ENST00000311487.9 |
HMGA1
|
high mobility group AT-hook 1 |
| chr3_+_41194741 | 0.17 |
ENST00000643541.1
ENST00000426215.5 ENST00000645210.1 ENST00000646381.1 ENST00000405570.6 ENST00000642248.1 ENST00000433400.6 |
CTNNB1
|
catenin beta 1 |
| chr1_-_110607425 | 0.17 |
ENST00000633222.1
|
KCNA2
|
potassium voltage-gated channel subfamily A member 2 |
| chr3_-_79019444 | 0.17 |
ENST00000618833.4
ENST00000436010.6 ENST00000618846.4 |
ROBO1
|
roundabout guidance receptor 1 |
| chr3_+_122055355 | 0.17 |
ENST00000330540.7
ENST00000469710.5 ENST00000493101.5 |
CD86
|
CD86 molecule |
| chr10_-_60572599 | 0.17 |
ENST00000503366.5
|
ANK3
|
ankyrin 3 |
| chr17_-_43546323 | 0.17 |
ENST00000545954.5
ENST00000319349.10 |
ETV4
|
ETS variant transcription factor 4 |
| chr2_-_196592671 | 0.17 |
ENST00000260983.8
ENST00000644030.1 |
HECW2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr3_+_171843337 | 0.16 |
ENST00000334567.9
ENST00000619900.4 ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr5_-_147453888 | 0.16 |
ENST00000398514.7
|
DPYSL3
|
dihydropyrimidinase like 3 |
| chr10_+_69801892 | 0.16 |
ENST00000398978.8
ENST00000645393.2 ENST00000354547.7 ENST00000674121.1 ENST00000673842.1 ENST00000520267.5 |
COL13A1
|
collagen type XIII alpha 1 chain |
| chr18_+_58341038 | 0.16 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr1_-_110607307 | 0.16 |
ENST00000639048.2
ENST00000675391.1 ENST00000639233.2 |
KCNA2
|
potassium voltage-gated channel subfamily A member 2 |
| chr3_-_24165548 | 0.15 |
ENST00000280696.9
|
THRB
|
thyroid hormone receptor beta |
| chr19_-_55738374 | 0.15 |
ENST00000590200.1
ENST00000332836.7 |
NLRP9
|
NLR family pyrin domain containing 9 |
| chr11_-_35419213 | 0.15 |
ENST00000642171.1
ENST00000644050.1 ENST00000643134.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr22_+_30080460 | 0.15 |
ENST00000336726.11
|
HORMAD2
|
HORMA domain containing 2 |
| chr1_-_959240 | 0.15 |
ENST00000327044.7
|
NOC2L
|
NOC2 like nucleolar associated transcriptional repressor |
| chr1_-_17439657 | 0.14 |
ENST00000375436.9
|
RCC2
|
regulator of chromosome condensation 2 |
| chr12_+_20695323 | 0.14 |
ENST00000266509.7
|
SLCO1C1
|
solute carrier organic anion transporter family member 1C1 |
| chr9_-_114657791 | 0.14 |
ENST00000423632.3
|
TEX53
|
testis expressed 53 |
| chr11_-_35418966 | 0.14 |
ENST00000531628.2
|
SLC1A2
|
solute carrier family 1 member 2 |
| chr6_+_57090069 | 0.14 |
ENST00000370708.8
ENST00000370702.5 |
ZNF451
|
zinc finger protein 451 |
| chr1_-_451678 | 0.14 |
ENST00000426406.4
|
OR4F29
|
olfactory receptor family 4 subfamily F member 29 |
| chr11_-_35419098 | 0.14 |
ENST00000606205.6
ENST00000645303.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr12_+_6724008 | 0.13 |
ENST00000626119.2
ENST00000543155.6 |
COPS7A
|
COP9 signalosome subunit 7A |
| chr7_+_120273129 | 0.13 |
ENST00000331113.9
|
KCND2
|
potassium voltage-gated channel subfamily D member 2 |
| chr3_-_71583713 | 0.13 |
ENST00000649528.3
ENST00000471386.3 ENST00000493089.7 |
FOXP1
|
forkhead box P1 |
| chr7_-_51316754 | 0.13 |
ENST00000632460.1
ENST00000441453.5 ENST00000648294.1 ENST00000265136.12 ENST00000395542.6 ENST00000395540.6 |
COBL
|
cordon-bleu WH2 repeat protein |
| chr11_-_116787979 | 0.13 |
ENST00000429220.5
ENST00000444935.5 ENST00000227322.8 |
ZPR1
|
ZPR1 zinc finger |
| chr3_+_69936629 | 0.13 |
ENST00000394348.2
ENST00000531774.1 |
MITF
|
melanocyte inducing transcription factor |
| chr15_+_66386902 | 0.12 |
ENST00000307102.10
|
MAP2K1
|
mitogen-activated protein kinase kinase 1 |
| chr1_-_159714581 | 0.12 |
ENST00000255030.9
ENST00000437342.1 ENST00000368112.5 ENST00000368111.5 ENST00000368110.1 |
CRP
|
C-reactive protein |
| chr7_-_23470469 | 0.12 |
ENST00000258729.8
|
IGF2BP3
|
insulin like growth factor 2 mRNA binding protein 3 |
| chr18_+_63542365 | 0.12 |
ENST00000269491.6
ENST00000382768.2 |
SERPINB12
|
serpin family B member 12 |
| chr12_-_4379712 | 0.12 |
ENST00000237837.2
|
FGF23
|
fibroblast growth factor 23 |
| chr9_+_100578071 | 0.12 |
ENST00000307584.6
|
CAVIN4
|
caveolae associated protein 4 |
| chr12_+_6724071 | 0.11 |
ENST00000229251.7
ENST00000539735.5 ENST00000538410.5 |
COPS7A
|
COP9 signalosome subunit 7A |
| chr3_-_186362223 | 0.11 |
ENST00000265022.8
|
DGKG
|
diacylglycerol kinase gamma |
| chr1_-_112618204 | 0.11 |
ENST00000369664.1
|
ST7L
|
suppression of tumorigenicity 7 like |
| chr16_+_56191728 | 0.11 |
ENST00000638705.1
ENST00000262494.12 |
GNAO1
|
G protein subunit alpha o1 |
| chr3_+_63443306 | 0.11 |
ENST00000472899.5
ENST00000479198.5 ENST00000460711.5 ENST00000465156.1 |
SYNPR
|
synaptoporin |
| chr3_-_171026709 | 0.11 |
ENST00000314251.8
|
SLC2A2
|
solute carrier family 2 member 2 |
| chr10_+_69802424 | 0.11 |
ENST00000673802.2
ENST00000517713.5 ENST00000520133.5 ENST00000522165.5 ENST00000673641.2 ENST00000673628.2 |
COL13A1
|
collagen type XIII alpha 1 chain |
| chr13_-_83882390 | 0.11 |
ENST00000377084.3
|
SLITRK1
|
SLIT and NTRK like family member 1 |
| chr22_-_35824373 | 0.11 |
ENST00000473487.6
|
RBFOX2
|
RNA binding fox-1 homolog 2 |
| chr11_-_107858777 | 0.10 |
ENST00000525815.6
|
SLC35F2
|
solute carrier family 35 member F2 |
| chr7_-_143362687 | 0.10 |
ENST00000409578.5
ENST00000443739.7 ENST00000409346.5 |
FAM131B
|
family with sequence similarity 131 member B |
| chr4_-_156970903 | 0.10 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr13_-_83882456 | 0.10 |
ENST00000674365.1
|
SLITRK1
|
SLIT and NTRK like family member 1 |
| chrY_-_17880220 | 0.10 |
ENST00000382867.4
|
CDY2B
|
chromodomain Y-linked 2B |
| chr4_-_68670648 | 0.10 |
ENST00000338206.6
|
UGT2B15
|
UDP glucuronosyltransferase family 2 member B15 |
| chr19_-_48513161 | 0.10 |
ENST00000673139.1
|
LMTK3
|
lemur tyrosine kinase 3 |
| chr9_-_27529705 | 0.10 |
ENST00000262244.6
|
MOB3B
|
MOB kinase activator 3B |
| chr3_-_71583683 | 0.10 |
ENST00000649631.1
ENST00000648718.1 |
FOXP1
|
forkhead box P1 |
| chr17_+_7558465 | 0.10 |
ENST00000349228.8
|
TNFSF13
|
TNF superfamily member 13 |
| chr19_-_52171600 | 0.10 |
ENST00000682614.1
|
ZNF836
|
zinc finger protein 836 |
| chr3_-_71583592 | 0.10 |
ENST00000650156.1
ENST00000649596.1 |
FOXP1
|
forkhead box P1 |
| chr1_-_29230871 | 0.10 |
ENST00000373791.7
ENST00000263702.11 |
MECR
|
mitochondrial trans-2-enoyl-CoA reductase |
| chr4_+_70592295 | 0.09 |
ENST00000449493.2
|
AMBN
|
ameloblastin |
| chr21_-_14546297 | 0.09 |
ENST00000400566.6
ENST00000400564.5 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr21_-_14546351 | 0.09 |
ENST00000619120.4
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr10_+_49637400 | 0.09 |
ENST00000640822.1
|
CHAT
|
choline O-acetyltransferase |
| chr3_-_155676363 | 0.09 |
ENST00000494598.5
|
PLCH1
|
phospholipase C eta 1 |
| chr8_+_119873710 | 0.09 |
ENST00000523492.5
ENST00000286234.6 |
DEPTOR
|
DEP domain containing MTOR interacting protein |
| chr14_+_22314435 | 0.09 |
ENST00000390467.3
|
TRAV40
|
T cell receptor alpha variable 40 |
| chr17_-_39607876 | 0.09 |
ENST00000302584.5
|
NEUROD2
|
neuronal differentiation 2 |
| chr1_+_149475933 | 0.09 |
ENST00000612881.4
|
NBPF19
|
NBPF member 19 |
| chr2_+_167868948 | 0.09 |
ENST00000392690.3
|
B3GALT1
|
beta-1,3-galactosyltransferase 1 |
| chr7_+_129368123 | 0.09 |
ENST00000460109.5
ENST00000474594.5 |
AHCYL2
|
adenosylhomocysteinase like 2 |
| chr1_-_115841116 | 0.09 |
ENST00000320238.3
|
NHLH2
|
nescient helix-loop-helix 2 |
| chr7_-_13986439 | 0.09 |
ENST00000443608.5
ENST00000438956.5 |
ETV1
|
ETS variant transcription factor 1 |
| chr12_-_91153149 | 0.09 |
ENST00000550758.1
|
DCN
|
decorin |
| chr17_-_41467386 | 0.08 |
ENST00000225899.4
|
KRT32
|
keratin 32 |
| chr15_+_92900189 | 0.08 |
ENST00000626874.2
ENST00000627622.1 ENST00000629346.2 ENST00000628375.2 ENST00000420239.7 ENST00000394196.9 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr5_-_156963222 | 0.08 |
ENST00000407087.4
ENST00000274532.7 |
TIMD4
|
T cell immunoglobulin and mucin domain containing 4 |
| chr6_-_138218491 | 0.08 |
ENST00000527246.3
|
PBOV1
|
prostate and breast cancer overexpressed 1 |
| chr6_-_75284820 | 0.08 |
ENST00000475111.6
|
TMEM30A
|
transmembrane protein 30A |
| chr14_-_23551729 | 0.08 |
ENST00000419474.5
|
ZFHX2
|
zinc finger homeobox 2 |
| chr1_-_31919093 | 0.08 |
ENST00000602683.5
ENST00000470404.5 |
PTP4A2
|
protein tyrosine phosphatase 4A2 |
| chr4_-_99290975 | 0.08 |
ENST00000209668.3
|
ADH1A
|
alcohol dehydrogenase 1A (class I), alpha polypeptide |
| chr4_-_110641920 | 0.08 |
ENST00000354925.6
ENST00000511990.1 ENST00000613094.4 ENST00000614423.4 ENST00000616641.4 ENST00000511837.5 |
PITX2
|
paired like homeodomain 2 |
| chr20_-_63956382 | 0.08 |
ENST00000358711.7
ENST00000354216.11 |
UCKL1
|
uridine-cytidine kinase 1 like 1 |
| chr12_+_48472559 | 0.08 |
ENST00000266594.4
|
ANP32D
|
acidic nuclear phosphoprotein 32 family member D |
| chr16_-_69754913 | 0.08 |
ENST00000268802.10
|
NOB1
|
NIN1 (RPN12) binding protein 1 homolog |
| chr17_-_69268812 | 0.08 |
ENST00000586811.1
|
ABCA5
|
ATP binding cassette subfamily A member 5 |
| chr3_-_11720728 | 0.07 |
ENST00000445411.5
ENST00000273038.7 |
VGLL4
|
vestigial like family member 4 |
| chr1_+_196977550 | 0.07 |
ENST00000256785.5
|
CFHR5
|
complement factor H related 5 |
| chr9_+_72114595 | 0.07 |
ENST00000545168.5
|
GDA
|
guanine deaminase |
| chr15_-_64381431 | 0.07 |
ENST00000558008.3
ENST00000300035.9 ENST00000559519.5 ENST00000380258.6 |
PCLAF
|
PCNA clamp associated factor |
| chr3_+_130931893 | 0.07 |
ENST00000504612.5
|
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr2_-_162838728 | 0.07 |
ENST00000328032.8
ENST00000332142.10 |
KCNH7
|
potassium voltage-gated channel subfamily H member 7 |
| chr3_-_120093666 | 0.07 |
ENST00000316626.6
ENST00000650344.2 ENST00000678439.1 |
GSK3B
|
glycogen synthase kinase 3 beta |
| chr5_+_127649018 | 0.07 |
ENST00000379445.7
|
CTXN3
|
cortexin 3 |
| chrY_+_18025787 | 0.07 |
ENST00000250838.6
|
CDY2A
|
chromodomain Y-linked 2A |
| chr19_-_52171643 | 0.06 |
ENST00000597065.1
|
ZNF836
|
zinc finger protein 836 |
| chr13_+_27792479 | 0.06 |
ENST00000302945.3
|
GSX1
|
GS homeobox 1 |
| chr19_-_4535221 | 0.06 |
ENST00000381848.7
ENST00000586133.1 |
PLIN5
|
perilipin 5 |
| chr15_+_92900338 | 0.06 |
ENST00000625990.3
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr8_-_144605699 | 0.06 |
ENST00000377307.6
ENST00000276826.5 |
ARHGAP39
|
Rho GTPase activating protein 39 |
| chr5_+_66144204 | 0.06 |
ENST00000612404.4
|
SREK1
|
splicing regulatory glutamic acid and lysine rich protein 1 |
| chr19_+_37506931 | 0.06 |
ENST00000627814.3
ENST00000587143.5 |
ZNF793
|
zinc finger protein 793 |
| chr2_-_162536955 | 0.06 |
ENST00000621889.1
|
KCNH7
|
potassium voltage-gated channel subfamily H member 7 |
| chr17_-_4263847 | 0.06 |
ENST00000570535.5
ENST00000574367.5 ENST00000341657.9 |
ANKFY1
|
ankyrin repeat and FYVE domain containing 1 |
| chr8_+_91249307 | 0.06 |
ENST00000309536.6
ENST00000276609.8 |
SLC26A7
|
solute carrier family 26 member 7 |
| chr12_+_41437680 | 0.06 |
ENST00000649474.1
ENST00000539469.6 ENST00000298919.7 |
PDZRN4
|
PDZ domain containing ring finger 4 |
| chr7_+_99828010 | 0.06 |
ENST00000631161.2
ENST00000354829.7 ENST00000342499.8 ENST00000417625.5 ENST00000415413.5 ENST00000444905.5 ENST00000222382.5 ENST00000312017.9 |
CYP3A43
|
cytochrome P450 family 3 subfamily A member 43 |
| chr2_-_206086057 | 0.06 |
ENST00000403263.6
|
INO80D
|
INO80 complex subunit D |
| chr3_-_142028597 | 0.06 |
ENST00000467667.5
|
TFDP2
|
transcription factor Dp-2 |
| chr4_+_157220691 | 0.06 |
ENST00000509417.5
ENST00000645636.1 ENST00000296526.12 ENST00000264426.14 |
GRIA2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr4_+_157220654 | 0.06 |
ENST00000393815.6
|
GRIA2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr9_-_135961310 | 0.05 |
ENST00000371756.4
|
UBAC1
|
UBA domain containing 1 |
| chr1_+_192636121 | 0.05 |
ENST00000543215.5
ENST00000391995.7 |
RGS13
|
regulator of G protein signaling 13 |
| chrX_+_91434829 | 0.05 |
ENST00000312600.4
|
PABPC5
|
poly(A) binding protein cytoplasmic 5 |
| chr14_+_69398683 | 0.05 |
ENST00000556605.5
ENST00000031146.8 ENST00000336643.10 |
SLC39A9
|
solute carrier family 39 member 9 |
| chr14_+_21797272 | 0.05 |
ENST00000390430.2
|
TRAV8-1
|
T cell receptor alpha variable 8-1 |
| chr12_+_64452084 | 0.05 |
ENST00000677641.1
ENST00000650790.1 ENST00000652657.1 ENST00000676912.1 ENST00000677545.1 ENST00000677632.1 ENST00000331710.10 ENST00000678180.1 ENST00000650762.1 ENST00000676930.1 ENST00000651014.1 ENST00000538890.5 |
TBK1
|
TANK binding kinase 1 |
| chr4_+_168497044 | 0.05 |
ENST00000505667.6
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr4_+_168497066 | 0.04 |
ENST00000261509.10
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr12_+_55362975 | 0.04 |
ENST00000641576.1
|
OR6C75
|
olfactory receptor family 6 subfamily C member 75 |
| chr9_+_137241277 | 0.04 |
ENST00000340384.5
|
TUBB4B
|
tubulin beta 4B class IVb |
| chr6_+_27957241 | 0.04 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor family 2 subfamily B member 6 |
| chr1_+_153217578 | 0.04 |
ENST00000368744.4
|
PRR9
|
proline rich 9 |
| chr19_+_7534004 | 0.04 |
ENST00000221249.10
ENST00000601668.5 ENST00000601001.5 |
PNPLA6
|
patatin like phospholipase domain containing 6 |
| chr20_+_37777262 | 0.04 |
ENST00000373469.1
|
CTNNBL1
|
catenin beta like 1 |
| chr6_-_41747390 | 0.04 |
ENST00000356667.8
ENST00000373025.7 ENST00000425343.6 |
PGC
|
progastricsin |
| chr16_-_8868343 | 0.04 |
ENST00000562843.5
ENST00000561530.5 ENST00000396593.6 |
CARHSP1
|
calcium regulated heat stable protein 1 |
| chr3_-_11643871 | 0.04 |
ENST00000430365.7
|
VGLL4
|
vestigial like family member 4 |
| chr9_-_96655280 | 0.04 |
ENST00000446045.1
ENST00000375234.8 |
PRXL2C
|
peroxiredoxin like 2C |
| chr6_+_31946086 | 0.04 |
ENST00000425368.7
|
CFB
|
complement factor B |
| chr3_+_130431463 | 0.04 |
ENST00000512836.5
|
COL6A5
|
collagen type VI alpha 5 chain |
| chr3_+_119782094 | 0.04 |
ENST00000393716.8
|
NR1I2
|
nuclear receptor subfamily 1 group I member 2 |
| chr7_+_142791635 | 0.04 |
ENST00000633705.1
|
TRBC1
|
T cell receptor beta constant 1 |
| chr17_+_81395449 | 0.04 |
ENST00000675386.2
|
BAHCC1
|
BAH domain and coiled-coil containing 1 |
| chr2_+_165239388 | 0.04 |
ENST00000424833.5
ENST00000375437.7 ENST00000631182.3 |
SCN2A
|
sodium voltage-gated channel alpha subunit 2 |
| chr10_-_119536533 | 0.04 |
ENST00000392865.5
|
RGS10
|
regulator of G protein signaling 10 |
| chr19_+_7534242 | 0.04 |
ENST00000545201.6
|
PNPLA6
|
patatin like phospholipase domain containing 6 |
| chr17_+_81395469 | 0.03 |
ENST00000584436.7
|
BAHCC1
|
BAH domain and coiled-coil containing 1 |
| chr10_-_96271553 | 0.03 |
ENST00000224337.10
|
BLNK
|
B cell linker |
| chr3_-_186544377 | 0.03 |
ENST00000307944.6
|
CRYGS
|
crystallin gamma S |
| chr12_+_56752449 | 0.03 |
ENST00000554643.5
ENST00000556650.5 ENST00000554150.5 ENST00000554155.1 |
HSD17B6
|
hydroxysteroid 17-beta dehydrogenase 6 |
| chr6_-_151452018 | 0.03 |
ENST00000491268.2
|
RMND1
|
required for meiotic nuclear division 1 homolog |
| chr5_-_157345645 | 0.03 |
ENST00000312349.5
|
FNDC9
|
fibronectin type III domain containing 9 |
| chr3_+_189171948 | 0.03 |
ENST00000345063.8
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr1_+_146949314 | 0.03 |
ENST00000617844.4
ENST00000611443.4 |
NBPF12
|
NBPF member 12 |
| chr1_+_92168915 | 0.03 |
ENST00000637221.2
|
BTBD8
|
BTB domain containing 8 |
| chr5_-_144170607 | 0.03 |
ENST00000448443.6
ENST00000513112.5 ENST00000519064.5 ENST00000274496.10 ENST00000522203.5 |
YIPF5
|
Yip1 domain family member 5 |
| chr17_+_38730301 | 0.03 |
ENST00000613478.2
|
CISD3
|
CDGSH iron sulfur domain 3 |
| chr2_+_170178136 | 0.03 |
ENST00000409044.7
ENST00000408978.9 |
MYO3B
|
myosin IIIB |
| chr16_-_20691256 | 0.03 |
ENST00000307493.8
|
ACSM1
|
acyl-CoA synthetase medium chain family member 1 |
| chr4_+_155903688 | 0.02 |
ENST00000536354.3
|
TDO2
|
tryptophan 2,3-dioxygenase |
| chrY_+_22403470 | 0.02 |
ENST00000250831.5
|
RBMY1J
|
RNA binding motif protein Y-linked family 1 member J |
| chr5_+_163460623 | 0.02 |
ENST00000393915.9
ENST00000432118.6 |
HMMR
|
hyaluronan mediated motility receptor |
| chr2_+_165239432 | 0.02 |
ENST00000636071.2
ENST00000636985.2 |
SCN2A
|
sodium voltage-gated channel alpha subunit 2 |
| chr16_-_8868687 | 0.02 |
ENST00000618335.4
ENST00000570125.5 |
CARHSP1
|
calcium regulated heat stable protein 1 |
| chr5_+_163460650 | 0.02 |
ENST00000358715.3
|
HMMR
|
hyaluronan mediated motility receptor |
| chr5_+_139561159 | 0.02 |
ENST00000505007.5
|
UBE2D2
|
ubiquitin conjugating enzyme E2 D2 |
| chrY_+_21511338 | 0.02 |
ENST00000383020.7
ENST00000619219.1 ENST00000439108.6 ENST00000303902.9 |
RBMY1B
RBMY1A1
|
RNA binding motif protein Y-linked family 1 member B RNA binding motif protein Y-linked family 1 member A1 |
| chr19_+_37507129 | 0.02 |
ENST00000586138.5
ENST00000588578.5 ENST00000587986.5 |
ZNF793
|
zinc finger protein 793 |
| chr20_+_59996335 | 0.02 |
ENST00000244049.7
ENST00000350849.10 ENST00000456106.1 |
CDH26
|
cadherin 26 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.1 | 0.9 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.1 | 0.2 | GO:0002668 | negative regulation of T cell tolerance induction(GO:0002665) negative regulation of T cell anergy(GO:0002668) negative regulation of lymphocyte anergy(GO:0002912) regulation of lymphotoxin A production(GO:0032681) positive regulation of lymphotoxin A production(GO:0032761) regulation of lymphotoxin A biosynthetic process(GO:0043016) positive regulation of lymphotoxin A biosynthetic process(GO:0043017) |
| 0.1 | 0.7 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.2 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.2 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.0 | 0.2 | GO:0021836 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.1 | GO:0071931 | positive regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071931) |
| 0.0 | 0.2 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.2 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.6 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.4 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:1904383 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) vitamin D catabolic process(GO:0042369) response to sodium phosphate(GO:1904383) |
| 0.0 | 0.2 | GO:1904793 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.0 | 0.4 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.2 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:0060578 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.0 | 0.1 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.1 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.2 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.3 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.1 | GO:2000077 | negative regulation of dopaminergic neuron differentiation(GO:1904339) negative regulation of type B pancreatic cell development(GO:2000077) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 0.0 | GO:1903423 | positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.1 | 0.2 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.1 | 0.9 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.1 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.0 | 0.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.2 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 1.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 1.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.7 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.4 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.4 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.0 | 0.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.8 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 1.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.7 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |