|
chr1_-_165445220
Show fit
|
0.86 |
ENST00000619224.1
|
RXRG
|
retinoid X receptor gamma
|
|
chr1_-_165445088
Show fit
|
0.86 |
ENST00000359842.10
|
RXRG
|
retinoid X receptor gamma
|
|
chr14_-_94770102
Show fit
|
0.80 |
ENST00000238558.5
|
GSC
|
goosecoid homeobox
|
|
chr8_+_53851786
Show fit
|
0.70 |
ENST00000297313.8
ENST00000344277.10
|
RGS20
|
regulator of G protein signaling 20
|
|
chr17_-_42112674
Show fit
|
0.49 |
ENST00000251642.8
ENST00000591220.5
|
DHX58
|
DExH-box helicase 58
|
|
chr22_-_37984534
Show fit
|
0.42 |
ENST00000396884.8
|
SOX10
|
SRY-box transcription factor 10
|
|
chr18_+_34710307
Show fit
|
0.40 |
ENST00000679796.1
|
DTNA
|
dystrobrevin alpha
|
|
chr11_-_62984690
Show fit
|
0.39 |
ENST00000421062.2
ENST00000458333.6
|
SLC22A6
|
solute carrier family 22 member 6
|
|
chr17_-_50129792
Show fit
|
0.37 |
ENST00000503131.1
|
SAMD14
|
sterile alpha motif domain containing 14
|
|
chr11_+_61481110
Show fit
|
0.36 |
ENST00000338608.7
ENST00000432063.6
|
PPP1R32
|
protein phosphatase 1 regulatory subunit 32
|
|
chr11_+_67455352
Show fit
|
0.35 |
ENST00000325656.7
|
CABP4
|
calcium binding protein 4
|
|
chr16_-_31064952
Show fit
|
0.34 |
ENST00000426488.6
|
ZNF668
|
zinc finger protein 668
|
|
chr16_-_31065011
Show fit
|
0.33 |
ENST00000539836.3
ENST00000535577.5
ENST00000442862.2
|
ZNF668
|
zinc finger protein 668
|
|
chr3_+_63967738
Show fit
|
0.32 |
ENST00000484332.1
|
ATXN7
|
ataxin 7
|
|
chrX_-_124963768
Show fit
|
0.31 |
ENST00000371130.7
ENST00000422452.2
|
TENM1
|
teneurin transmembrane protein 1
|
|
chr12_+_77830886
Show fit
|
0.31 |
ENST00000397909.7
ENST00000549464.5
|
NAV3
|
neuron navigator 3
|
|
chr11_+_20022550
Show fit
|
0.30 |
ENST00000533917.5
|
NAV2
|
neuron navigator 2
|
|
chr19_+_45498439
Show fit
|
0.28 |
ENST00000451287.7
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent 1N (putative)
|
|
chr2_+_120013111
Show fit
|
0.26 |
ENST00000331393.8
ENST00000443124.5
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5
|
|
chr11_-_62984957
Show fit
|
0.26 |
ENST00000377871.7
ENST00000360421.9
|
SLC22A6
|
solute carrier family 22 member 6
|
|
chr5_-_150289941
Show fit
|
0.26 |
ENST00000682786.1
|
CAMK2A
|
calcium/calmodulin dependent protein kinase II alpha
|
|
chr6_-_42722590
Show fit
|
0.25 |
ENST00000230381.7
|
PRPH2
|
peripherin 2
|
|
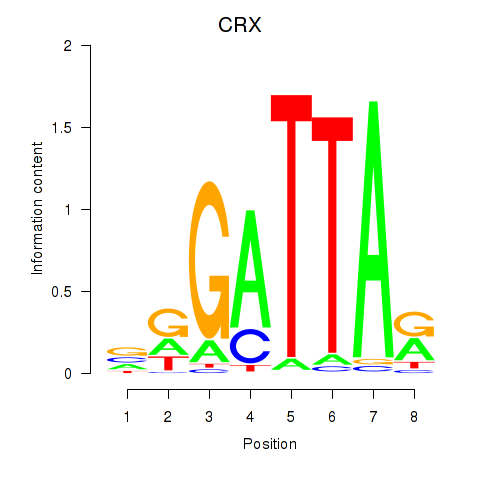
chr19_+_47821907
Show fit
|
0.25 |
ENST00000539067.5
ENST00000221996.12
ENST00000613299.1
|
CRX
|
cone-rod homeobox
|
|
chr16_+_57976435
Show fit
|
0.25 |
ENST00000290871.10
ENST00000441824.4
|
TEPP
|
testis, prostate and placenta expressed
|
|
chr8_-_132111159
Show fit
|
0.25 |
ENST00000673615.1
ENST00000434736.6
|
HHLA1
|
HERV-H LTR-associating 1
|
|
chr17_-_9905248
Show fit
|
0.23 |
ENST00000226193.6
|
RCVRN
|
recoverin
|
|
chr10_-_84241538
Show fit
|
0.22 |
ENST00000372105.4
|
LRIT1
|
leucine rich repeat, Ig-like and transmembrane domains 1
|
|
chr17_-_40703744
Show fit
|
0.22 |
ENST00000264651.3
|
KRT24
|
keratin 24
|
|
chr5_+_162067458
Show fit
|
0.20 |
ENST00000639975.1
ENST00000639111.2
ENST00000639683.1
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2
|
|
chr4_+_89111521
Show fit
|
0.19 |
ENST00000603357.3
|
TIGD2
|
tigger transposable element derived 2
|
|
chr18_+_34710249
Show fit
|
0.19 |
ENST00000680346.1
ENST00000348997.9
ENST00000681274.1
ENST00000680822.1
ENST00000680767.2
ENST00000597599.5
ENST00000444659.6
|
DTNA
|
dystrobrevin alpha
|
|
chr3_-_112499358
Show fit
|
0.19 |
ENST00000383680.4
|
BTLA
|
B and T lymphocyte associated
|
|
chr3_-_54928044
Show fit
|
0.18 |
ENST00000273286.6
|
LRTM1
|
leucine rich repeats and transmembrane domains 1
|
|
chr14_-_63318933
Show fit
|
0.17 |
ENST00000621500.2
|
GPHB5
|
glycoprotein hormone subunit beta 5
|
|
chr5_+_162067764
Show fit
|
0.17 |
ENST00000639213.2
ENST00000414552.6
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2
|
|
chr5_+_38258373
Show fit
|
0.17 |
ENST00000354891.7
|
EGFLAM
|
EGF like, fibronectin type III and laminin G domains
|
|
chr14_+_88005128
Show fit
|
0.16 |
ENST00000267549.5
|
GPR65
|
G protein-coupled receptor 65
|
|
chr12_+_57522801
Show fit
|
0.16 |
ENST00000355673.8
ENST00000546632.1
ENST00000549623.1
|
MBD6
|
methyl-CpG binding domain protein 6
|
|
chr3_-_112499457
Show fit
|
0.15 |
ENST00000334529.10
|
BTLA
|
B and T lymphocyte associated
|
|
chr13_-_33205997
Show fit
|
0.15 |
ENST00000399365.7
|
STARD13
|
StAR related lipid transfer domain containing 13
|
|
chr2_+_169509693
Show fit
|
0.14 |
ENST00000284669.2
|
KLHL41
|
kelch like family member 41
|
|
chr1_+_205257180
Show fit
|
0.14 |
ENST00000330675.12
|
TMCC2
|
transmembrane and coiled-coil domain family 2
|
|
chr12_+_80707625
Show fit
|
0.14 |
ENST00000228641.4
|
MYF6
|
myogenic factor 6
|
|
chr5_-_115626161
Show fit
|
0.14 |
ENST00000282382.8
|
TMED7-TICAM2
|
TMED7-TICAM2 readthrough
|
|
chr6_+_31587268
Show fit
|
0.14 |
ENST00000396101.7
ENST00000490742.5
|
LST1
|
leukocyte specific transcript 1
|
|
chr11_-_128842467
Show fit
|
0.14 |
ENST00000392664.2
|
KCNJ1
|
potassium inwardly rectifying channel subfamily J member 1
|
|
chr13_-_94479671
Show fit
|
0.14 |
ENST00000377028.10
ENST00000446125.1
|
DCT
|
dopachrome tautomerase
|
|
chr2_-_174847015
Show fit
|
0.14 |
ENST00000650938.1
|
CHN1
|
chimerin 1
|
|
chr5_+_136058849
Show fit
|
0.14 |
ENST00000508076.5
|
TGFBI
|
transforming growth factor beta induced
|
|
chr2_-_61538180
Show fit
|
0.14 |
ENST00000677150.1
ENST00000678182.1
ENST00000677928.1
ENST00000406957.5
|
XPO1
|
exportin 1
|
|
chr9_+_69145463
Show fit
|
0.14 |
ENST00000636438.1
|
TJP2
|
tight junction protein 2
|
|
chr10_+_102226293
Show fit
|
0.14 |
ENST00000370005.4
|
ELOVL3
|
ELOVL fatty acid elongase 3
|
|
chr1_+_18107763
Show fit
|
0.13 |
ENST00000251296.4
|
IGSF21
|
immunoglobin superfamily member 21
|
|
chr2_-_174846405
Show fit
|
0.13 |
ENST00000409597.5
ENST00000413882.6
|
CHN1
|
chimerin 1
|
|
chr7_-_78771265
Show fit
|
0.13 |
ENST00000630991.2
ENST00000629359.2
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr5_-_150289625
Show fit
|
0.13 |
ENST00000683332.1
ENST00000398376.8
ENST00000672785.1
ENST00000672396.1
|
CAMK2A
|
calcium/calmodulin dependent protein kinase II alpha
|
|
chr2_-_61538290
Show fit
|
0.13 |
ENST00000678790.1
|
XPO1
|
exportin 1
|
|
chr18_-_66604076
Show fit
|
0.13 |
ENST00000540086.5
ENST00000580157.2
ENST00000262150.7
|
CDH19
|
cadherin 19
|
|
chr5_-_36301883
Show fit
|
0.13 |
ENST00000502994.5
ENST00000515759.5
ENST00000296604.8
|
RANBP3L
|
RAN binding protein 3 like
|
|
chr3_+_111674654
Show fit
|
0.13 |
ENST00000636933.1
ENST00000393934.7
ENST00000477665.2
|
PLCXD2
|
phosphatidylinositol specific phospholipase C X domain containing 2
|
|
chr3_-_186544377
Show fit
|
0.12 |
ENST00000307944.6
|
CRYGS
|
crystallin gamma S
|
|
chr3_+_1092654
Show fit
|
0.12 |
ENST00000350110.2
|
CNTN6
|
contactin 6
|
|
chr12_-_56454653
Show fit
|
0.12 |
ENST00000652304.1
|
MIP
|
major intrinsic protein of lens fiber
|
|
chr3_+_1093002
Show fit
|
0.12 |
ENST00000446702.7
|
CNTN6
|
contactin 6
|
|
chr15_+_58138368
Show fit
|
0.12 |
ENST00000219919.9
ENST00000536493.1
|
AQP9
|
aquaporin 9
|
|
chr4_+_128809791
Show fit
|
0.12 |
ENST00000452328.6
ENST00000504089.5
|
JADE1
|
jade family PHD finger 1
|
|
chr19_-_3772211
Show fit
|
0.12 |
ENST00000555978.5
ENST00000555633.3
|
RAX2
|
retina and anterior neural fold homeobox 2
|
|
chr3_+_173584433
Show fit
|
0.12 |
ENST00000361589.8
|
NLGN1
|
neuroligin 1
|
|
chr15_+_86079863
Show fit
|
0.11 |
ENST00000614907.3
ENST00000441037.7
|
AGBL1
|
ATP/GTP binding protein like 1
|
|
chr11_+_96389985
Show fit
|
0.11 |
ENST00000332349.5
|
JRKL
|
JRK like
|
|
chr5_+_162067500
Show fit
|
0.11 |
ENST00000639384.1
ENST00000640985.1
ENST00000638772.1
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2
|
|
chr6_+_31587185
Show fit
|
0.11 |
ENST00000376092.7
ENST00000376086.7
ENST00000303757.12
ENST00000376093.6
|
LST1
|
leukocyte specific transcript 1
|
|
chrX_-_32412220
Show fit
|
0.11 |
ENST00000619831.5
|
DMD
|
dystrophin
|
|
chr1_-_117121692
Show fit
|
0.11 |
ENST00000256649.9
ENST00000369464.7
ENST00000485032.1
|
TRIM45
|
tripartite motif containing 45
|
|
chr2_-_61537740
Show fit
|
0.11 |
ENST00000678081.1
ENST00000676889.1
ENST00000677850.1
ENST00000676789.1
|
XPO1
|
exportin 1
|
|
chrX_+_15749848
Show fit
|
0.11 |
ENST00000479740.5
ENST00000454127.2
|
CA5B
|
carbonic anhydrase 5B
|
|
chr4_-_152679984
Show fit
|
0.11 |
ENST00000304385.8
ENST00000504064.1
|
TMEM154
|
transmembrane protein 154
|
|
chr2_-_24047348
Show fit
|
0.11 |
ENST00000406895.3
|
WDCP
|
WD repeat and coiled coil containing
|
|
chr2_-_24047375
Show fit
|
0.11 |
ENST00000295148.9
|
WDCP
|
WD repeat and coiled coil containing
|
|
chr5_+_64165815
Show fit
|
0.10 |
ENST00000389100.9
ENST00000296615.10
|
RNF180
|
ring finger protein 180
|
|
chr2_-_144431001
Show fit
|
0.10 |
ENST00000636413.1
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr15_+_79311137
Show fit
|
0.10 |
ENST00000424155.6
ENST00000536821.5
|
TMED3
|
transmembrane p24 trafficking protein 3
|
|
chr12_+_14973020
Show fit
|
0.10 |
ENST00000266395.3
|
PDE6H
|
phosphodiesterase 6H
|
|
chr11_+_124865425
Show fit
|
0.10 |
ENST00000397801.6
|
ROBO3
|
roundabout guidance receptor 3
|
|
chr6_+_35342535
Show fit
|
0.10 |
ENST00000360694.8
ENST00000418635.6
ENST00000448077.6
|
PPARD
|
peroxisome proliferator activated receptor delta
|
|
chr19_-_42427379
Show fit
|
0.10 |
ENST00000244289.9
|
LIPE
|
lipase E, hormone sensitive type
|
|
chr10_-_92243246
Show fit
|
0.10 |
ENST00000412050.8
ENST00000614585.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr19_-_48044037
Show fit
|
0.10 |
ENST00000293255.3
|
CABP5
|
calcium binding protein 5
|
|
chr7_-_78771108
Show fit
|
0.10 |
ENST00000626691.2
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr9_-_115118198
Show fit
|
0.10 |
ENST00000534839.1
ENST00000535648.5
|
TNC
|
tenascin C
|
|
chr12_+_104303726
Show fit
|
0.10 |
ENST00000527879.2
|
EID3
|
EP300 interacting inhibitor of differentiation 3
|
|
chr3_-_190862688
Show fit
|
0.10 |
ENST00000442080.6
|
GMNC
|
geminin coiled-coil domain containing
|
|
chr18_-_72865680
Show fit
|
0.09 |
ENST00000397929.5
|
NETO1
|
neuropilin and tolloid like 1
|
|
chr18_+_616672
Show fit
|
0.09 |
ENST00000338387.11
|
CLUL1
|
clusterin like 1
|
|
chr17_+_8002610
Show fit
|
0.09 |
ENST00000254854.5
|
GUCY2D
|
guanylate cyclase 2D, retinal
|
|
chr11_-_30586866
Show fit
|
0.09 |
ENST00000528686.2
|
MPPED2
|
metallophosphoesterase domain containing 2
|
|
chr18_+_616711
Show fit
|
0.09 |
ENST00000579494.1
|
CLUL1
|
clusterin like 1
|
|
chr6_-_138572523
Show fit
|
0.09 |
ENST00000427025.6
|
NHSL1
|
NHS like 1
|
|
chr3_+_129528632
Show fit
|
0.09 |
ENST00000296271.4
|
RHO
|
rhodopsin
|
|
chr5_+_162067858
Show fit
|
0.09 |
ENST00000361925.9
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2
|
|
chr11_-_67523396
Show fit
|
0.09 |
ENST00000353903.9
ENST00000294288.5
|
CABP2
|
calcium binding protein 2
|
|
chr9_-_128204218
Show fit
|
0.09 |
ENST00000634901.1
ENST00000372948.7
|
CIZ1
|
CDKN1A interacting zinc finger protein 1
|
|
chr16_-_57971121
Show fit
|
0.09 |
ENST00000251102.13
|
CNGB1
|
cyclic nucleotide gated channel subunit beta 1
|
|
chr4_-_110636963
Show fit
|
0.09 |
ENST00000394595.8
|
PITX2
|
paired like homeodomain 2
|
|
chr6_+_54308429
Show fit
|
0.09 |
ENST00000259782.9
ENST00000370864.3
|
TINAG
|
tubulointerstitial nephritis antigen
|
|
chr20_+_44355692
Show fit
|
0.09 |
ENST00000316673.8
ENST00000609795.5
ENST00000457232.5
ENST00000609262.5
|
HNF4A
|
hepatocyte nuclear factor 4 alpha
|
|
chr12_+_7789393
Show fit
|
0.09 |
ENST00000229307.9
|
NANOG
|
Nanog homeobox
|
|
chr4_+_153222307
Show fit
|
0.08 |
ENST00000675899.1
ENST00000675611.1
ENST00000674872.1
ENST00000676167.1
|
TRIM2
|
tripartite motif containing 2
|
|
chr4_-_99563984
Show fit
|
0.08 |
ENST00000394877.7
|
TRMT10A
|
tRNA methyltransferase 10A
|
|
chr6_+_12716760
Show fit
|
0.08 |
ENST00000332995.12
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr2_-_70553440
Show fit
|
0.08 |
ENST00000450929.5
|
TGFA
|
transforming growth factor alpha
|
|
chr5_-_149944744
Show fit
|
0.08 |
ENST00000255266.10
ENST00000617647.4
ENST00000613228.1
|
PDE6A
|
phosphodiesterase 6A
|
|
chr4_+_128809684
Show fit
|
0.08 |
ENST00000226319.11
ENST00000511647.5
|
JADE1
|
jade family PHD finger 1
|
|
chr19_+_11346499
Show fit
|
0.08 |
ENST00000458408.6
ENST00000586451.5
ENST00000588592.5
|
CCDC159
|
coiled-coil domain containing 159
|
|
chr9_+_18474100
Show fit
|
0.08 |
ENST00000327883.11
ENST00000431052.6
ENST00000380570.8
ENST00000380548.9
|
ADAMTSL1
|
ADAMTS like 1
|
|
chr6_+_31587049
Show fit
|
0.08 |
ENST00000376089.6
ENST00000396112.6
|
LST1
|
leukocyte specific transcript 1
|
|
chr12_+_133181409
Show fit
|
0.08 |
ENST00000416488.5
ENST00000228289.9
ENST00000541211.6
ENST00000536435.7
ENST00000500625.7
ENST00000539248.6
ENST00000542711.6
ENST00000536899.6
ENST00000542986.6
ENST00000611984.4
ENST00000541975.2
|
ZNF268
|
zinc finger protein 268
|
|
chr21_+_34668986
Show fit
|
0.08 |
ENST00000349499.3
|
CLIC6
|
chloride intracellular channel 6
|
|
chr4_-_99563668
Show fit
|
0.08 |
ENST00000273962.7
|
TRMT10A
|
tRNA methyltransferase 10A
|
|
chr3_-_48188356
Show fit
|
0.08 |
ENST00000351231.7
ENST00000437972.1
ENST00000302506.8
|
CDC25A
|
cell division cycle 25A
|
|
chr9_-_92482350
Show fit
|
0.08 |
ENST00000375543.2
|
ASPN
|
asporin
|
|
chr3_-_50611767
Show fit
|
0.08 |
ENST00000443053.6
ENST00000348721.4
|
CISH
|
cytokine inducible SH2 containing protein
|
|
chr4_+_153222402
Show fit
|
0.08 |
ENST00000676335.1
ENST00000675146.1
|
TRIM2
|
tripartite motif containing 2
|
|
chr9_-_92482499
Show fit
|
0.08 |
ENST00000375544.7
|
ASPN
|
asporin
|
|
chr1_+_244051275
Show fit
|
0.08 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18
|
|
chr5_-_24644968
Show fit
|
0.07 |
ENST00000264463.8
|
CDH10
|
cadherin 10
|
|
chrX_+_140091415
Show fit
|
0.07 |
ENST00000453380.2
|
ENSG00000230707.2
|
novel protein (LOC389895)
|
|
chr2_-_65432591
Show fit
|
0.07 |
ENST00000356388.9
|
SPRED2
|
sprouty related EVH1 domain containing 2
|
|
chr11_+_102112445
Show fit
|
0.07 |
ENST00000524575.5
|
YAP1
|
Yes1 associated transcriptional regulator
|
|
chr11_-_41459592
Show fit
|
0.07 |
ENST00000528697.6
ENST00000530763.5
|
LRRC4C
|
leucine rich repeat containing 4C
|
|
chr6_+_130018565
Show fit
|
0.07 |
ENST00000361794.7
ENST00000526087.5
ENST00000533560.5
|
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3
|
|
chr9_+_34179005
Show fit
|
0.07 |
ENST00000625521.2
ENST00000379186.8
ENST00000297661.9
ENST00000626262.2
|
UBAP1
|
ubiquitin associated protein 1
|
|
chr7_+_80602150
Show fit
|
0.07 |
ENST00000309881.11
|
CD36
|
CD36 molecule
|
|
chr7_+_80602200
Show fit
|
0.07 |
ENST00000534394.5
|
CD36
|
CD36 molecule
|
|
chr1_-_201112420
Show fit
|
0.07 |
ENST00000362061.4
ENST00000681874.1
|
CACNA1S
|
calcium voltage-gated channel subunit alpha1 S
|
|
chr1_+_167329044
Show fit
|
0.07 |
ENST00000367862.9
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr10_-_13972355
Show fit
|
0.07 |
ENST00000264546.10
|
FRMD4A
|
FERM domain containing 4A
|
|
chr11_-_47176851
Show fit
|
0.07 |
ENST00000629231.2
ENST00000526342.5
ENST00000528444.5
ENST00000530596.5
ENST00000525398.5
ENST00000524782.6
ENST00000527927.5
ENST00000525314.5
|
ARFGAP2
|
ADP ribosylation factor GTPase activating protein 2
|
|
chr11_-_126062782
Show fit
|
0.07 |
ENST00000531738.6
|
CDON
|
cell adhesion associated, oncogene regulated
|
|
chr2_+_66435558
Show fit
|
0.07 |
ENST00000488550.5
|
MEIS1
|
Meis homeobox 1
|
|
chr11_-_129024157
Show fit
|
0.06 |
ENST00000392657.7
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr15_-_65115185
Show fit
|
0.06 |
ENST00000559089.6
|
UBAP1L
|
ubiquitin associated protein 1 like
|
|
chr19_+_39498938
Show fit
|
0.06 |
ENST00000356433.10
ENST00000596614.5
ENST00000205143.4
|
DLL3
|
delta like canonical Notch ligand 3
|
|
chr7_+_21428023
Show fit
|
0.06 |
ENST00000432066.2
ENST00000222584.8
|
SP4
|
Sp4 transcription factor
|
|
chr2_-_174634566
Show fit
|
0.06 |
ENST00000392547.6
|
WIPF1
|
WAS/WASL interacting protein family member 1
|
|
chr6_+_31586680
Show fit
|
0.06 |
ENST00000339530.8
|
LST1
|
leukocyte specific transcript 1
|
|
chr7_-_84194781
Show fit
|
0.06 |
ENST00000265362.9
|
SEMA3A
|
semaphorin 3A
|
|
chr10_-_87818153
Show fit
|
0.06 |
ENST00000308448.11
ENST00000680024.1
|
ATAD1
|
ATPase family AAA domain containing 1
|
|
chr1_-_11691608
Show fit
|
0.06 |
ENST00000376667.7
|
MAD2L2
|
mitotic arrest deficient 2 like 2
|
|
chr3_+_130931893
Show fit
|
0.06 |
ENST00000504612.5
|
ATP2C1
|
ATPase secretory pathway Ca2+ transporting 1
|
|
chr5_-_524322
Show fit
|
0.06 |
ENST00000644203.1
ENST00000514375.1
ENST00000264938.8
|
SLC9A3
|
solute carrier family 9 member A3
|
|
chr12_-_91182652
Show fit
|
0.06 |
ENST00000552145.5
ENST00000546745.5
|
DCN
|
decorin
|
|
chr1_-_11691646
Show fit
|
0.06 |
ENST00000235310.7
|
MAD2L2
|
mitotic arrest deficient 2 like 2
|
|
chrX_-_21658324
Show fit
|
0.06 |
ENST00000379499.3
|
KLHL34
|
kelch like family member 34
|
|
chr12_-_118190510
Show fit
|
0.06 |
ENST00000540561.5
ENST00000537952.1
ENST00000537822.1
|
TAOK3
|
TAO kinase 3
|
|
chr1_+_11691688
Show fit
|
0.06 |
ENST00000294485.6
|
DRAXIN
|
dorsal inhibitory axon guidance protein
|
|
chr4_+_70028452
Show fit
|
0.06 |
ENST00000530128.5
ENST00000381057.3
ENST00000673563.1
|
HTN3
|
histatin 3
|
|
chr1_+_55039511
Show fit
|
0.06 |
ENST00000302118.5
|
PCSK9
|
proprotein convertase subtilisin/kexin type 9
|
|
chr11_-_118565810
Show fit
|
0.06 |
ENST00000672656.2
ENST00000530872.5
ENST00000264021.8
|
IFT46
|
intraflagellar transport 46
|
|
chr6_+_99606833
Show fit
|
0.06 |
ENST00000369215.5
|
PRDM13
|
PR/SET domain 13
|
|
chr9_-_28670285
Show fit
|
0.06 |
ENST00000379992.6
ENST00000308675.5
ENST00000613945.3
|
LINGO2
|
leucine rich repeat and Ig domain containing 2
|
|
chr14_-_36320582
Show fit
|
0.06 |
ENST00000604336.5
ENST00000359527.11
ENST00000621657.4
ENST00000603139.1
ENST00000318473.11
ENST00000416007.9
|
MBIP
|
MAP3K12 binding inhibitory protein 1
|
|
chr7_+_90709231
Show fit
|
0.05 |
ENST00000446790.5
ENST00000265741.7
|
CDK14
|
cyclin dependent kinase 14
|
|
chr2_+_183078736
Show fit
|
0.05 |
ENST00000354221.5
|
DUSP19
|
dual specificity phosphatase 19
|
|
chr8_-_25424260
Show fit
|
0.05 |
ENST00000421054.7
|
GNRH1
|
gonadotropin releasing hormone 1
|
|
chr1_-_11691471
Show fit
|
0.05 |
ENST00000376672.5
|
MAD2L2
|
mitotic arrest deficient 2 like 2
|
|
chr15_+_79311084
Show fit
|
0.05 |
ENST00000299705.10
|
TMED3
|
transmembrane p24 trafficking protein 3
|
|
chr7_-_98252117
Show fit
|
0.05 |
ENST00000420697.1
ENST00000415086.5
ENST00000447648.7
|
TECPR1
|
tectonin beta-propeller repeat containing 1
|
|
chr11_-_118565992
Show fit
|
0.05 |
ENST00000264020.6
|
IFT46
|
intraflagellar transport 46
|
|
chr1_+_159171607
Show fit
|
0.05 |
ENST00000368124.8
ENST00000368125.9
ENST00000416746.1
|
CADM3
|
cell adhesion molecule 3
|
|
chr19_-_43619591
Show fit
|
0.05 |
ENST00000598676.1
ENST00000300811.8
|
ZNF428
|
zinc finger protein 428
|
|
chr2_+_66435116
Show fit
|
0.05 |
ENST00000272369.14
ENST00000560281.6
|
MEIS1
|
Meis homeobox 1
|
|
chr4_+_153204410
Show fit
|
0.05 |
ENST00000675838.1
ENST00000674967.1
ENST00000632856.2
ENST00000441616.6
ENST00000433687.2
ENST00000494872.6
ENST00000460908.2
ENST00000675780.1
ENST00000674976.1
ENST00000338700.10
ENST00000675293.1
ENST00000676172.1
ENST00000675673.1
ENST00000675492.1
ENST00000675425.1
ENST00000675384.1
ENST00000675063.1
ENST00000675340.1
ENST00000675835.1
ENST00000675054.1
ENST00000675710.1
ENST00000502281.3
|
ENSG00000288637.1
TRIM2
|
novel protein
tripartite motif containing 2
|
|
chr17_+_77281429
Show fit
|
0.05 |
ENST00000591198.5
ENST00000427177.6
|
SEPTIN9
|
septin 9
|
|
chr5_+_173889337
Show fit
|
0.05 |
ENST00000520867.5
ENST00000334035.9
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4
|
|
chr3_+_4493340
Show fit
|
0.05 |
ENST00000357086.10
ENST00000354582.12
ENST00000649015.2
ENST00000467056.6
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor type 1
|
|
chr6_+_30326835
Show fit
|
0.05 |
ENST00000440271.5
ENST00000376656.8
ENST00000396551.7
ENST00000428728.5
ENST00000396548.5
ENST00000428404.5
|
TRIM39
|
tripartite motif containing 39
|
|
chr21_-_18403754
Show fit
|
0.05 |
ENST00000284885.8
|
TMPRSS15
|
transmembrane serine protease 15
|
|
chr10_-_75109172
Show fit
|
0.05 |
ENST00000372700.7
ENST00000473072.2
ENST00000491677.6
ENST00000372702.7
|
DUSP13
|
dual specificity phosphatase 13
|
|
chr10_-_32378720
Show fit
|
0.05 |
ENST00000375110.6
|
EPC1
|
enhancer of polycomb homolog 1
|
|
chr17_+_45241067
Show fit
|
0.05 |
ENST00000587489.5
|
FMNL1
|
formin like 1
|
|
chr2_+_191276885
Show fit
|
0.05 |
ENST00000392316.5
|
MYO1B
|
myosin IB
|
|
chr7_+_74453790
Show fit
|
0.05 |
ENST00000265755.7
ENST00000424337.7
ENST00000455841.6
|
GTF2IRD1
|
GTF2I repeat domain containing 1
|
|
chr3_-_146528750
Show fit
|
0.04 |
ENST00000483300.5
|
PLSCR1
|
phospholipid scramblase 1
|
|
chr16_-_57971086
Show fit
|
0.04 |
ENST00000564448.5
ENST00000311183.8
|
CNGB1
|
cyclic nucleotide gated channel subunit beta 1
|
|
chr5_-_11588842
Show fit
|
0.04 |
ENST00000503622.5
|
CTNND2
|
catenin delta 2
|
|
chr10_+_22321056
Show fit
|
0.04 |
ENST00000376663.8
|
BMI1
|
BMI1 proto-oncogene, polycomb ring finger
|
|
chr1_+_26317950
Show fit
|
0.04 |
ENST00000374213.3
|
CD52
|
CD52 molecule
|
|
chr2_-_212124901
Show fit
|
0.04 |
ENST00000402597.6
|
ERBB4
|
erb-b2 receptor tyrosine kinase 4
|
|
chr15_-_74202742
Show fit
|
0.04 |
ENST00000395105.9
|
STRA6
|
signaling receptor and transporter of retinol STRA6
|
|
chr15_-_31160954
Show fit
|
0.04 |
ENST00000558445.5
ENST00000559177.5
|
TRPM1
|
transient receptor potential cation channel subfamily M member 1
|
|
chr12_+_53299682
Show fit
|
0.04 |
ENST00000267103.10
ENST00000548632.5
|
MYG1
|
MYG1 exonuclease
|
|
chr11_-_36598221
Show fit
|
0.04 |
ENST00000311485.8
ENST00000527033.5
ENST00000532616.1
ENST00000618712.4
|
RAG2
|
recombination activating 2
|
|
chr5_-_96433214
Show fit
|
0.04 |
ENST00000311106.8
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr2_-_224497816
Show fit
|
0.04 |
ENST00000451538.1
|
CUL3
|
cullin 3
|
|
chr11_-_119346695
Show fit
|
0.04 |
ENST00000619721.6
|
MFRP
|
membrane frizzled-related protein
|
|
chrM_+_9207
Show fit
|
0.04 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III
|
|
chr10_+_102132994
Show fit
|
0.04 |
ENST00000413464.6
ENST00000278070.7
|
PPRC1
|
PPARG related coactivator 1
|
|
chr5_-_150758497
Show fit
|
0.04 |
ENST00000521533.1
ENST00000424236.5
|
DCTN4
|
dynactin subunit 4
|
|
chr10_+_94089034
Show fit
|
0.04 |
ENST00000676102.1
ENST00000371385.8
|
PLCE1
|
phospholipase C epsilon 1
|