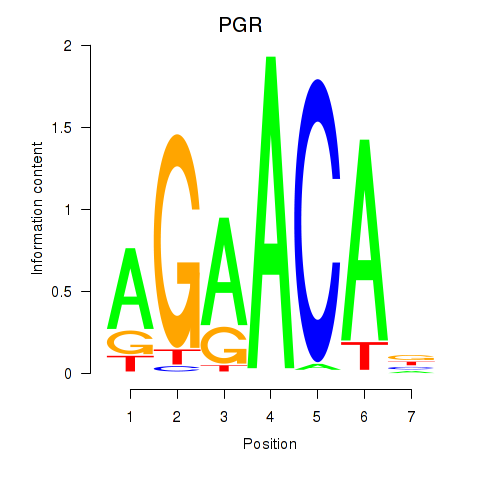
|
chr3_-_149333407
Show fit
|
1.84 |
ENST00000470080.5
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr17_-_44199206
Show fit
|
1.66 |
ENST00000589805.1
|
ATXN7L3
|
ataxin 7 like 3
|
|
chr12_+_78864768
Show fit
|
1.62 |
ENST00000261205.9
ENST00000457153.6
|
SYT1
|
synaptotagmin 1
|
|
chr9_-_104928139
Show fit
|
1.45 |
ENST00000423487.6
ENST00000374733.1
ENST00000374736.8
ENST00000678995.1
|
ABCA1
|
ATP binding cassette subfamily A member 1
|
|
chr1_+_148748774
Show fit
|
1.34 |
ENST00000322209.5
|
NUDT4B
|
nudix hydrolase 4B
|
|
chr6_-_75493629
Show fit
|
1.30 |
ENST00000393004.6
|
FILIP1
|
filamin A interacting protein 1
|
|
chr1_+_25616780
Show fit
|
1.30 |
ENST00000374332.9
|
MAN1C1
|
mannosidase alpha class 1C member 1
|
|
chr4_-_185775271
Show fit
|
1.29 |
ENST00000430503.5
ENST00000319454.10
ENST00000450341.5
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr2_+_172821575
Show fit
|
1.29 |
ENST00000397087.7
|
RAPGEF4
|
Rap guanine nucleotide exchange factor 4
|
|
chr2_-_187448244
Show fit
|
1.20 |
ENST00000392370.8
ENST00000410068.5
ENST00000447403.5
ENST00000410102.5
|
CALCRL
|
calcitonin receptor like receptor
|
|
chr5_-_95081482
Show fit
|
1.14 |
ENST00000312216.12
ENST00000512425.5
ENST00000505208.5
ENST00000429576.6
ENST00000508509.5
ENST00000510732.5
|
MCTP1
|
multiple C2 and transmembrane domain containing 1
|
|
chr4_-_185775890
Show fit
|
1.12 |
ENST00000437304.6
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr6_-_75493773
Show fit
|
1.08 |
ENST00000237172.12
|
FILIP1
|
filamin A interacting protein 1
|
|
chr3_-_149333619
Show fit
|
1.07 |
ENST00000296059.7
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr4_-_89838289
Show fit
|
1.03 |
ENST00000336904.7
|
SNCA
|
synuclein alpha
|
|
chr15_+_31366138
Show fit
|
1.00 |
ENST00000558844.1
|
KLF13
|
Kruppel like factor 13
|
|
chr12_+_78863962
Show fit
|
0.99 |
ENST00000393240.7
|
SYT1
|
synaptotagmin 1
|
|
chr18_+_44700796
Show fit
|
0.98 |
ENST00000677130.1
|
SETBP1
|
SET binding protein 1
|
|
chr6_-_75206044
Show fit
|
0.85 |
ENST00000322507.13
|
COL12A1
|
collagen type XII alpha 1 chain
|
|
chr22_-_28712136
Show fit
|
0.82 |
ENST00000464581.6
|
CHEK2
|
checkpoint kinase 2
|
|
chr6_-_36839434
Show fit
|
0.76 |
ENST00000244751.7
ENST00000633280.1
|
CPNE5
|
copine 5
|
|
chr5_-_88731827
Show fit
|
0.72 |
ENST00000627170.2
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr11_+_6876625
Show fit
|
0.71 |
ENST00000379829.2
|
OR10A4
|
olfactory receptor family 10 subfamily A member 4
|
|
chr16_-_10559135
Show fit
|
0.70 |
ENST00000536829.1
|
EMP2
|
epithelial membrane protein 2
|
|
chr9_+_2159672
Show fit
|
0.69 |
ENST00000634343.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2
|
|
chr12_+_7061206
Show fit
|
0.67 |
ENST00000423384.5
ENST00000413211.5
|
C1S
|
complement C1s
|
|
chr20_+_36091409
Show fit
|
0.62 |
ENST00000202028.9
|
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1
|
|
chr3_+_52420955
Show fit
|
0.61 |
ENST00000465863.1
|
PHF7
|
PHD finger protein 7
|
|
chr6_+_73695779
Show fit
|
0.61 |
ENST00000422508.6
ENST00000437994.6
|
CD109
|
CD109 molecule
|
|
chr9_+_113463697
Show fit
|
0.58 |
ENST00000317613.10
|
RGS3
|
regulator of G protein signaling 3
|
|
chr14_-_34713759
Show fit
|
0.58 |
ENST00000673315.1
|
CFL2
|
cofilin 2
|
|
chr8_-_30083053
Show fit
|
0.57 |
ENST00000256255.11
ENST00000523761.1
|
SARAF
|
store-operated calcium entry associated regulatory factor
|
|
chr8_-_30083110
Show fit
|
0.57 |
ENST00000545648.2
|
SARAF
|
store-operated calcium entry associated regulatory factor
|
|
chr9_+_2159850
Show fit
|
0.57 |
ENST00000416751.2
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2
|
|
chrX_-_45200895
Show fit
|
0.57 |
ENST00000377934.4
|
DIPK2B
|
divergent protein kinase domain 2B
|
|
chrX_-_101386166
Show fit
|
0.57 |
ENST00000308731.8
ENST00000372880.5
|
BTK
|
Bruton tyrosine kinase
|
|
chr1_-_85404494
Show fit
|
0.56 |
ENST00000633113.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chrX_+_54807599
Show fit
|
0.55 |
ENST00000375053.6
ENST00000627068.2
ENST00000375068.6
ENST00000347546.8
|
MAGED2
|
MAGE family member D2
|
|
chr2_-_182522703
Show fit
|
0.55 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A
|
|
chr18_-_55586092
Show fit
|
0.55 |
ENST00000563888.6
ENST00000540999.5
ENST00000627685.2
|
TCF4
|
transcription factor 4
|
|
chr2_-_215436061
Show fit
|
0.55 |
ENST00000421182.5
ENST00000432072.6
ENST00000323926.10
ENST00000336916.8
ENST00000357867.8
ENST00000359671.5
ENST00000446046.5
ENST00000354785.11
ENST00000356005.8
ENST00000443816.5
ENST00000426059.1
|
FN1
|
fibronectin 1
|
|
chr20_-_23086316
Show fit
|
0.55 |
ENST00000246006.5
|
CD93
|
CD93 molecule
|
|
chr7_+_130486171
Show fit
|
0.53 |
ENST00000341441.9
ENST00000416162.7
|
MEST
|
mesoderm specific transcript
|
|
chr19_+_30372364
Show fit
|
0.51 |
ENST00000355537.4
|
ZNF536
|
zinc finger protein 536
|
|
chr10_+_47322450
Show fit
|
0.51 |
ENST00000581492.3
|
GDF2
|
growth differentiation factor 2
|
|
chr20_-_25339731
Show fit
|
0.49 |
ENST00000450393.5
ENST00000491682.5
|
ABHD12
|
abhydrolase domain containing 12, lysophospholipase
|
|
chr8_+_22589240
Show fit
|
0.48 |
ENST00000450780.6
ENST00000430850.6
ENST00000447849.2
ENST00000614502.4
ENST00000443561.3
|
ENSG00000248235.6
PDLIM2
|
novel protein
PDZ and LIM domain 2
|
|
chr5_+_141417659
Show fit
|
0.48 |
ENST00000398594.4
|
PCDHGB7
|
protocadherin gamma subfamily B, 7
|
|
chr9_-_15510954
Show fit
|
0.48 |
ENST00000380733.9
|
PSIP1
|
PC4 and SFRS1 interacting protein 1
|
|
chr6_-_46954922
Show fit
|
0.47 |
ENST00000265417.7
|
ADGRF5
|
adhesion G protein-coupled receptor F5
|
|
chr12_-_71157992
Show fit
|
0.45 |
ENST00000247829.8
|
TSPAN8
|
tetraspanin 8
|
|
chr2_-_182242031
Show fit
|
0.45 |
ENST00000358139.6
|
PDE1A
|
phosphodiesterase 1A
|
|
chr1_+_92168915
Show fit
|
0.45 |
ENST00000637221.2
|
BTBD8
|
BTB domain containing 8
|
|
chr18_-_55351977
Show fit
|
0.43 |
ENST00000643689.1
|
TCF4
|
transcription factor 4
|
|
chr1_-_110391041
Show fit
|
0.43 |
ENST00000369781.8
ENST00000437429.6
ENST00000541986.5
|
SLC16A4
|
solute carrier family 16 member 4
|
|
chr12_-_6606427
Show fit
|
0.43 |
ENST00000642879.1
|
CHD4
|
chromodomain helicase DNA binding protein 4
|
|
chr12_-_71157872
Show fit
|
0.42 |
ENST00000546561.2
|
TSPAN8
|
tetraspanin 8
|
|
chrX_-_45200828
Show fit
|
0.42 |
ENST00000398000.7
|
DIPK2B
|
divergent protein kinase domain 2B
|
|
chrX_+_101488044
Show fit
|
0.42 |
ENST00000423738.4
|
ARMCX4
|
armadillo repeat containing X-linked 4
|
|
chr6_+_73696145
Show fit
|
0.41 |
ENST00000287097.6
|
CD109
|
CD109 molecule
|
|
chr12_-_6606642
Show fit
|
0.40 |
ENST00000545584.2
ENST00000545942.6
|
CHD4
|
chromodomain helicase DNA binding protein 4
|
|
chr2_-_74440484
Show fit
|
0.40 |
ENST00000305557.9
ENST00000233330.6
|
RTKN
|
rhotekin
|
|
chr9_+_113536497
Show fit
|
0.40 |
ENST00000462143.5
|
RGS3
|
regulator of G protein signaling 3
|
|
chrX_-_112840815
Show fit
|
0.39 |
ENST00000304758.5
ENST00000371959.9
|
AMOT
|
angiomotin
|
|
chr19_+_51693327
Show fit
|
0.38 |
ENST00000637797.2
|
SPACA6
|
sperm acrosome associated 6
|
|
chr12_-_6606320
Show fit
|
0.38 |
ENST00000642594.1
ENST00000644289.1
ENST00000645095.1
|
CHD4
|
chromodomain helicase DNA binding protein 4
|
|
chr13_-_99016034
Show fit
|
0.38 |
ENST00000448493.7
|
DOCK9
|
dedicator of cytokinesis 9
|
|
chr7_+_143959927
Show fit
|
0.37 |
ENST00000624504.1
|
OR2F1
|
olfactory receptor family 2 subfamily F member 1
|
|
chr19_+_13024917
Show fit
|
0.37 |
ENST00000587260.1
|
NFIX
|
nuclear factor I X
|
|
chr17_+_76001715
Show fit
|
0.36 |
ENST00000586261.2
|
CDK3
|
cyclin dependent kinase 3
|
|
chr3_-_114624193
Show fit
|
0.36 |
ENST00000481632.5
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chrX_+_15507302
Show fit
|
0.35 |
ENST00000342014.6
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr6_-_154510675
Show fit
|
0.35 |
ENST00000607772.6
|
CNKSR3
|
CNKSR family member 3
|
|
chr9_-_15510991
Show fit
|
0.34 |
ENST00000380715.5
ENST00000380716.8
ENST00000380738.8
|
PSIP1
|
PC4 and SFRS1 interacting protein 1
|
|
chr17_-_7857139
Show fit
|
0.34 |
ENST00000576384.1
|
NAA38
|
N-alpha-acetyltransferase 38, NatC auxiliary subunit
|
|
chr17_-_44389594
Show fit
|
0.34 |
ENST00000262407.6
|
ITGA2B
|
integrin subunit alpha 2b
|
|
chr14_+_65411845
Show fit
|
0.34 |
ENST00000556518.5
ENST00000557164.5
|
FUT8
|
fucosyltransferase 8
|
|
chr16_-_28495519
Show fit
|
0.33 |
ENST00000569430.7
|
CLN3
|
CLN3 lysosomal/endosomal transmembrane protein, battenin
|
|
chr14_-_34713788
Show fit
|
0.33 |
ENST00000341223.8
|
CFL2
|
cofilin 2
|
|
chr17_-_73092657
Show fit
|
0.33 |
ENST00000580557.5
ENST00000579732.5
ENST00000578620.1
ENST00000542342.6
ENST00000255559.7
ENST00000579018.5
|
SLC39A11
|
solute carrier family 39 member 11
|
|
chr5_+_175871670
Show fit
|
0.33 |
ENST00000514150.5
|
CPLX2
|
complexin 2
|
|
chr17_+_19282979
Show fit
|
0.33 |
ENST00000395626.5
ENST00000571254.1
|
EPN2
|
epsin 2
|
|
chr2_-_55334529
Show fit
|
0.33 |
ENST00000645860.1
ENST00000642563.1
ENST00000647396.1
|
CCDC88A
|
coiled-coil domain containing 88A
|
|
chr8_+_81732434
Show fit
|
0.32 |
ENST00000297265.5
|
CHMP4C
|
charged multivesicular body protein 4C
|
|
chr1_-_155991198
Show fit
|
0.32 |
ENST00000673475.1
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor 2
|
|
chr2_+_190343561
Show fit
|
0.32 |
ENST00000322522.8
ENST00000392329.7
ENST00000430311.5
|
INPP1
|
inositol polyphosphate-1-phosphatase
|
|
chrX_+_151694967
Show fit
|
0.32 |
ENST00000448726.5
ENST00000538575.5
|
PRRG3
|
proline rich and Gla domain 3
|
|
chr17_-_7857461
Show fit
|
0.31 |
ENST00000575771.6
ENST00000575071.5
|
NAA38
|
N-alpha-acetyltransferase 38, NatC auxiliary subunit
|
|
chr3_+_148865288
Show fit
|
0.31 |
ENST00000296046.4
|
CPA3
|
carboxypeptidase A3
|
|
chr6_+_31655888
Show fit
|
0.31 |
ENST00000375916.4
|
APOM
|
apolipoprotein M
|
|
chr3_+_25428233
Show fit
|
0.29 |
ENST00000437042.6
ENST00000330688.9
|
RARB
|
retinoic acid receptor beta
|
|
chr14_+_24171853
Show fit
|
0.29 |
ENST00000620473.4
ENST00000557806.5
ENST00000611366.5
|
REC8
|
REC8 meiotic recombination protein
|
|
chr2_+_162318884
Show fit
|
0.28 |
ENST00000446271.5
ENST00000429691.6
|
GCA
|
grancalcin
|
|
chr8_-_7430348
Show fit
|
0.28 |
ENST00000318124.3
|
DEFB103B
|
defensin beta 103B
|
|
chr7_-_93226449
Show fit
|
0.28 |
ENST00000394468.7
ENST00000453812.2
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr15_+_65550819
Show fit
|
0.28 |
ENST00000569894.5
|
HACD3
|
3-hydroxyacyl-CoA dehydratase 3
|
|
chr6_-_159045010
Show fit
|
0.28 |
ENST00000338313.5
|
TAGAP
|
T cell activation RhoGTPase activating protein
|
|
chr8_+_132919403
Show fit
|
0.28 |
ENST00000519178.5
|
TG
|
thyroglobulin
|
|
chr1_-_205321737
Show fit
|
0.28 |
ENST00000367157.6
|
NUAK2
|
NUAK family kinase 2
|
|
chr5_+_139293660
Show fit
|
0.27 |
ENST00000512876.5
ENST00000513678.5
|
MATR3
|
matrin 3
|
|
chr15_+_73683938
Show fit
|
0.26 |
ENST00000567189.5
|
CD276
|
CD276 molecule
|
|
chr20_-_61998132
Show fit
|
0.26 |
ENST00000474089.5
|
TAF4
|
TATA-box binding protein associated factor 4
|
|
chr10_-_77140757
Show fit
|
0.25 |
ENST00000637862.2
|
KCNMA1
|
potassium calcium-activated channel subfamily M alpha 1
|
|
chr8_+_10095704
Show fit
|
0.25 |
ENST00000382490.9
|
MSRA
|
methionine sulfoxide reductase A
|
|
chr7_-_130441136
Show fit
|
0.25 |
ENST00000675596.1
ENST00000676312.1
|
CEP41
|
centrosomal protein 41
|
|
chr2_+_10420021
Show fit
|
0.24 |
ENST00000422133.1
|
HPCAL1
|
hippocalcin like 1
|
|
chr14_-_67488709
Show fit
|
0.24 |
ENST00000554480.6
|
TMEM229B
|
transmembrane protein 229B
|
|
chr14_-_21476874
Show fit
|
0.24 |
ENST00000649801.1
ENST00000397762.6
|
RAB2B
|
RAB2B, member RAS oncogene family
|
|
chr14_+_22516273
Show fit
|
0.24 |
ENST00000390510.1
|
TRAJ27
|
T cell receptor alpha joining 27
|
|
chr19_+_48874318
Show fit
|
0.24 |
ENST00000600406.1
|
PPP1R15A
|
protein phosphatase 1 regulatory subunit 15A
|
|
chr12_+_57455266
Show fit
|
0.24 |
ENST00000266646.3
|
INHBE
|
inhibin subunit beta E
|
|
chr11_-_73982830
Show fit
|
0.24 |
ENST00000536983.5
ENST00000663595.2
ENST00000310473.9
|
UCP2
|
uncoupling protein 2
|
|
chr5_+_102865805
Show fit
|
0.23 |
ENST00000346918.7
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr5_-_135034212
Show fit
|
0.23 |
ENST00000265340.12
|
PITX1
|
paired like homeodomain 1
|
|
chr11_+_6481473
Show fit
|
0.23 |
ENST00000530751.1
ENST00000254616.11
|
TIMM10B
|
translocase of inner mitochondrial membrane 10B
|
|
chr12_-_108731505
Show fit
|
0.23 |
ENST00000261401.8
ENST00000552871.5
|
CORO1C
|
coronin 1C
|
|
chr11_-_5441514
Show fit
|
0.22 |
ENST00000380211.1
|
OR51I1
|
olfactory receptor family 51 subfamily I member 1
|
|
chr3_-_114624921
Show fit
|
0.22 |
ENST00000393785.6
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr17_+_47209338
Show fit
|
0.22 |
ENST00000393450.5
|
MYL4
|
myosin light chain 4
|
|
chr5_+_66828762
Show fit
|
0.22 |
ENST00000490016.6
ENST00000403666.5
ENST00000450827.5
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr19_-_49361475
Show fit
|
0.22 |
ENST00000598810.5
|
TEAD2
|
TEA domain transcription factor 2
|
|
chr6_-_32190170
Show fit
|
0.22 |
ENST00000375050.6
|
PBX2
|
PBX homeobox 2
|
|
chr3_-_185821092
Show fit
|
0.22 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2
|
|
chrX_+_154429092
Show fit
|
0.22 |
ENST00000619046.5
|
ATP6AP1
|
ATPase H+ transporting accessory protein 1
|
|
chr11_-_113706330
Show fit
|
0.21 |
ENST00000540540.5
ENST00000538955.5
ENST00000545579.6
|
TMPRSS5
|
transmembrane serine protease 5
|
|
chr17_+_47209035
Show fit
|
0.21 |
ENST00000572316.5
ENST00000354968.5
ENST00000576874.5
ENST00000536623.6
|
MYL4
|
myosin light chain 4
|
|
chr3_+_111542134
Show fit
|
0.21 |
ENST00000438817.6
|
CD96
|
CD96 molecule
|
|
chr5_+_139293939
Show fit
|
0.21 |
ENST00000508689.1
ENST00000514528.5
|
MATR3
|
matrin 3
|
|
chr2_-_157874976
Show fit
|
0.21 |
ENST00000682025.1
ENST00000683487.1
ENST00000682300.1
ENST00000683441.1
ENST00000684595.1
ENST00000683426.1
ENST00000683820.1
ENST00000263640.7
|
ACVR1
|
activin A receptor type 1
|
|
chr2_+_96326204
Show fit
|
0.21 |
ENST00000420728.1
ENST00000361124.5
|
ITPRIPL1
|
ITPRIP like 1
|
|
chr5_-_38557459
Show fit
|
0.20 |
ENST00000511561.1
|
LIFR
|
LIF receptor subunit alpha
|
|
chr18_+_63777773
Show fit
|
0.20 |
ENST00000447428.5
ENST00000546027.5
|
SERPINB7
|
serpin family B member 7
|
|
chr17_+_47209375
Show fit
|
0.20 |
ENST00000572303.1
|
MYL4
|
myosin light chain 4
|
|
chr2_-_27308445
Show fit
|
0.20 |
ENST00000296099.2
|
UCN
|
urocortin
|
|
chr12_+_8914464
Show fit
|
0.20 |
ENST00000544916.6
|
PHC1
|
polyhomeotic homolog 1
|
|
chr5_+_179820895
Show fit
|
0.20 |
ENST00000504627.1
ENST00000389805.9
ENST00000510187.5
|
SQSTM1
|
sequestosome 1
|
|
chr10_-_70382589
Show fit
|
0.20 |
ENST00000373224.5
ENST00000446961.4
ENST00000357631.6
ENST00000358141.6
ENST00000395010.5
|
LRRC20
|
leucine rich repeat containing 20
|
|
chr7_+_74093079
Show fit
|
0.20 |
ENST00000538333.3
|
LIMK1
|
LIM domain kinase 1
|
|
chrX_+_65588368
Show fit
|
0.20 |
ENST00000609672.5
|
MSN
|
moesin
|
|
chr5_-_64768619
Show fit
|
0.20 |
ENST00000513458.9
|
SREK1IP1
|
SREK1 interacting protein 1
|
|
chr8_-_101790934
Show fit
|
0.20 |
ENST00000523645.5
ENST00000520346.1
ENST00000220931.11
ENST00000522448.5
ENST00000522951.5
ENST00000522252.5
ENST00000519098.5
|
NCALD
|
neurocalcin delta
|
|
chr3_+_42091316
Show fit
|
0.20 |
ENST00000327628.10
|
TRAK1
|
trafficking kinesin protein 1
|
|
chr8_+_40153475
Show fit
|
0.19 |
ENST00000315792.5
|
TCIM
|
transcriptional and immune response regulator
|
|
chr1_+_152719522
Show fit
|
0.19 |
ENST00000368775.3
|
C1orf68
|
chromosome 1 open reading frame 68
|
|
chr13_+_73058993
Show fit
|
0.19 |
ENST00000377687.6
|
KLF5
|
Kruppel like factor 5
|
|
chr7_+_74027770
Show fit
|
0.19 |
ENST00000445912.5
ENST00000621115.4
|
ELN
|
elastin
|
|
chr10_+_122271292
Show fit
|
0.19 |
ENST00000260723.6
|
BTBD16
|
BTB domain containing 16
|
|
chr1_-_183590876
Show fit
|
0.19 |
ENST00000367536.5
|
NCF2
|
neutrophil cytosolic factor 2
|
|
chr12_-_48788995
Show fit
|
0.19 |
ENST00000550422.5
ENST00000357869.8
|
ADCY6
|
adenylate cyclase 6
|
|
chr2_+_225399684
Show fit
|
0.19 |
ENST00000636099.1
|
NYAP2
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2
|
|
chr4_-_151015713
Show fit
|
0.19 |
ENST00000357115.9
|
LRBA
|
LPS responsive beige-like anchor protein
|
|
chr6_+_20401864
Show fit
|
0.19 |
ENST00000346618.8
ENST00000613242.4
|
E2F3
|
E2F transcription factor 3
|
|
chr15_-_43584864
Show fit
|
0.18 |
ENST00000381885.5
ENST00000396923.7
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1
|
|
chr3_+_159273235
Show fit
|
0.18 |
ENST00000638749.1
|
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough
|
|
chr5_+_70025534
Show fit
|
0.18 |
ENST00000515588.1
|
SERF1B
|
small EDRK-rich factor 1B
|
|
chr8_+_10095551
Show fit
|
0.18 |
ENST00000522907.5
ENST00000528246.5
|
MSRA
|
methionine sulfoxide reductase A
|
|
chr22_-_40856565
Show fit
|
0.18 |
ENST00000620312.4
ENST00000216218.8
|
ST13
|
ST13 Hsp70 interacting protein
|
|
chr4_+_127880876
Show fit
|
0.18 |
ENST00000270861.10
ENST00000515069.5
ENST00000513090.5
ENST00000507249.5
|
PLK4
|
polo like kinase 4
|
|
chr17_-_4560564
Show fit
|
0.18 |
ENST00000574584.1
ENST00000381550.8
ENST00000301395.7
|
GGT6
|
gamma-glutamyltransferase 6
|
|
chr12_+_56267674
Show fit
|
0.18 |
ENST00000546544.5
ENST00000553234.1
|
COQ10A
|
coenzyme Q10A
|
|
chr11_-_625168
Show fit
|
0.18 |
ENST00000349570.11
|
CDHR5
|
cadherin related family member 5
|
|
chr7_+_74028127
Show fit
|
0.18 |
ENST00000438880.5
ENST00000414324.5
ENST00000380562.8
ENST00000380575.8
ENST00000380584.8
ENST00000458204.5
ENST00000357036.9
ENST00000417091.5
ENST00000429192.5
ENST00000252034.12
ENST00000442310.5
ENST00000380553.8
ENST00000380576.9
ENST00000428787.5
ENST00000320399.10
|
ELN
|
elastin
|
|
chr14_-_21580076
Show fit
|
0.18 |
ENST00000641040.1
ENST00000641185.1
|
OR10G3
|
olfactory receptor family 10 subfamily G member 3
|
|
chr17_+_48892761
Show fit
|
0.18 |
ENST00000355938.9
ENST00000393366.7
ENST00000503641.5
ENST00000514808.5
ENST00000506855.1
|
ATP5MC1
|
ATP synthase membrane subunit c locus 1
|
|
chr11_-_113706292
Show fit
|
0.17 |
ENST00000544634.5
ENST00000539732.5
ENST00000538770.1
ENST00000536856.5
ENST00000299882.11
ENST00000544476.1
|
TMPRSS5
|
transmembrane serine protease 5
|
|
chr7_+_23598144
Show fit
|
0.17 |
ENST00000410069.1
|
CCDC126
|
coiled-coil domain containing 126
|
|
chr17_-_35943662
Show fit
|
0.17 |
ENST00000618542.4
|
LYZL6
|
lysozyme like 6
|
|
chr17_+_35809474
Show fit
|
0.17 |
ENST00000604879.5
ENST00000603427.5
ENST00000603777.5
ENST00000605844.6
ENST00000604841.5
|
TAF15
|
TATA-box binding protein associated factor 15
|
|
chr7_+_22727147
Show fit
|
0.17 |
ENST00000426291.5
ENST00000401651.5
ENST00000258743.10
ENST00000407492.5
ENST00000401630.7
ENST00000406575.1
|
IL6
|
interleukin 6
|
|
chr13_+_101452629
Show fit
|
0.17 |
ENST00000622834.4
ENST00000545560.6
ENST00000376180.8
|
ITGBL1
|
integrin subunit beta like 1
|
|
chr10_+_113125536
Show fit
|
0.17 |
ENST00000349937.7
|
TCF7L2
|
transcription factor 7 like 2
|
|
chr17_-_35943707
Show fit
|
0.17 |
ENST00000615905.5
|
LYZL6
|
lysozyme like 6
|
|
chr3_-_114624979
Show fit
|
0.17 |
ENST00000676079.1
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr1_-_32336224
Show fit
|
0.17 |
ENST00000329421.8
|
MARCKSL1
|
MARCKS like 1
|
|
chr3_-_151384741
Show fit
|
0.16 |
ENST00000302632.4
|
P2RY12
|
purinergic receptor P2Y12
|
|
chr4_-_57110373
Show fit
|
0.16 |
ENST00000295666.6
ENST00000514062.2
|
IGFBP7
|
insulin like growth factor binding protein 7
|
|
chrX_-_79367307
Show fit
|
0.16 |
ENST00000373298.7
|
ITM2A
|
integral membrane protein 2A
|
|
chrX_+_16946862
Show fit
|
0.15 |
ENST00000303843.7
|
REPS2
|
RALBP1 associated Eps domain containing 2
|
|
chr1_-_26067622
Show fit
|
0.15 |
ENST00000374272.4
|
TRIM63
|
tripartite motif containing 63
|
|
chr17_-_3433841
Show fit
|
0.15 |
ENST00000248384.1
|
OR1E2
|
olfactory receptor family 1 subfamily E member 2
|
|
chr9_+_124853417
Show fit
|
0.15 |
ENST00000613760.4
ENST00000618744.4
ENST00000373574.2
|
WDR38
|
WD repeat domain 38
|
|
chr9_+_126326809
Show fit
|
0.15 |
ENST00000361171.8
ENST00000489637.3
|
MVB12B
|
multivesicular body subunit 12B
|
|
chr18_+_58864866
Show fit
|
0.14 |
ENST00000588456.5
ENST00000591808.6
ENST00000589481.1
ENST00000591049.1
|
ZNF532
|
zinc finger protein 532
|
|
chr4_-_138242325
Show fit
|
0.14 |
ENST00000280612.9
|
SLC7A11
|
solute carrier family 7 member 11
|
|
chr16_-_3256587
Show fit
|
0.14 |
ENST00000536379.5
ENST00000541159.5
ENST00000339854.8
ENST00000219596.6
|
MEFV
|
MEFV innate immuity regulator, pyrin
|
|
chr19_+_35282520
Show fit
|
0.14 |
ENST00000222304.5
|
HAMP
|
hepcidin antimicrobial peptide
|
|
chr19_-_55179390
Show fit
|
0.14 |
ENST00000590851.5
|
SYT5
|
synaptotagmin 5
|
|
chr2_+_48568981
Show fit
|
0.13 |
ENST00000394754.5
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough
|
|
chr6_+_39792298
Show fit
|
0.13 |
ENST00000633794.1
ENST00000274867.9
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chrY_+_14522573
Show fit
|
0.13 |
ENST00000643089.1
ENST00000382872.5
|
NLGN4Y
|
neuroligin 4 Y-linked
|
|
chrX_+_41447322
Show fit
|
0.13 |
ENST00000378220.2
ENST00000342595.2
|
NYX
|
nyctalopin
|
|
chr10_+_62049211
Show fit
|
0.13 |
ENST00000309334.5
|
ARID5B
|
AT-rich interaction domain 5B
|
|
chr6_-_43308797
Show fit
|
0.13 |
ENST00000274990.4
ENST00000372569.8
|
CRIP3
|
cysteine rich protein 3
|
|
chr15_+_92904447
Show fit
|
0.12 |
ENST00000626782.2
|
CHD2
|
chromodomain helicase DNA binding protein 2
|
|
chr22_-_31292445
Show fit
|
0.12 |
ENST00000402249.7
ENST00000215912.10
ENST00000443175.1
ENST00000441972.5
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1
|
|
chr10_-_73252601
Show fit
|
0.12 |
ENST00000372945.8
ENST00000372940.3
|
MRPS16
|
mitochondrial ribosomal protein S16
|
|
chrX_+_57592011
Show fit
|
0.12 |
ENST00000374888.3
|
ZXDB
|
zinc finger X-linked duplicated B
|
|
chr8_+_10672646
Show fit
|
0.12 |
ENST00000521818.1
|
C8orf74
|
chromosome 8 open reading frame 74
|
|
chr19_+_51127030
Show fit
|
0.12 |
ENST00000599948.1
|
SIGLEC9
|
sialic acid binding Ig like lectin 9
|
|
chr8_+_406808
Show fit
|
0.12 |
ENST00000352684.2
|
FBXO25
|
F-box protein 25
|