Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for PITX2
Z-value: 0.52
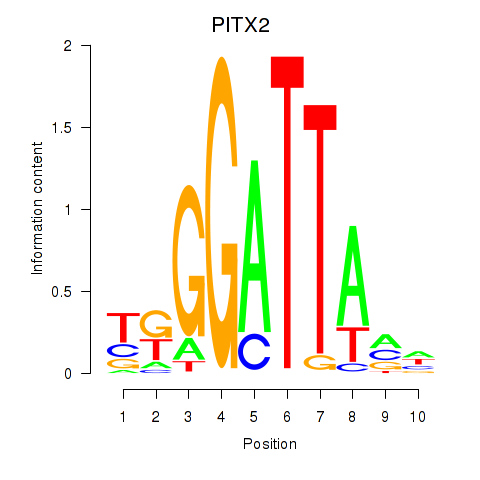
Transcription factors associated with PITX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PITX2
|
ENSG00000164093.17 | PITX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PITX2 | hg38_v1_chr4_-_110623051_110623085, hg38_v1_chr4_-_110636963_110637042, hg38_v1_chr4_-_110641920_110642123 | -0.41 | 4.5e-02 | Click! |
Activity profile of PITX2 motif
Sorted Z-values of PITX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PITX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_2476118 | 2.07 |
ENST00000215631.9
ENST00000587345.1 |
GADD45B
|
growth arrest and DNA damage inducible beta |
| chr9_-_14300231 | 1.27 |
ENST00000636735.1
|
NFIB
|
nuclear factor I B |
| chr4_-_101347492 | 1.23 |
ENST00000394854.8
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr4_-_101347327 | 1.05 |
ENST00000394853.8
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr4_-_101347471 | 1.01 |
ENST00000323055.10
ENST00000512215.5 |
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr4_-_101346842 | 1.00 |
ENST00000507176.5
|
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr6_-_111483700 | 0.83 |
ENST00000435970.5
ENST00000358835.7 |
REV3L
|
REV3 like, DNA directed polymerase zeta catalytic subunit |
| chr15_+_40929338 | 0.79 |
ENST00000249749.7
|
DLL4
|
delta like canonical Notch ligand 4 |
| chr17_+_4740005 | 0.77 |
ENST00000269289.10
|
ZMYND15
|
zinc finger MYND-type containing 15 |
| chr17_+_4740042 | 0.77 |
ENST00000592813.5
|
ZMYND15
|
zinc finger MYND-type containing 15 |
| chr17_-_41518878 | 0.62 |
ENST00000254043.8
|
KRT15
|
keratin 15 |
| chr11_-_119346655 | 0.61 |
ENST00000360167.4
|
MFRP
|
membrane frizzled-related protein |
| chr6_+_35342535 | 0.61 |
ENST00000360694.8
ENST00000418635.6 ENST00000448077.6 |
PPARD
|
peroxisome proliferator activated receptor delta |
| chr1_+_43300971 | 0.59 |
ENST00000372476.8
ENST00000538015.1 |
TIE1
|
tyrosine kinase with immunoglobulin like and EGF like domains 1 |
| chr6_+_130018565 | 0.52 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chr12_-_54981838 | 0.49 |
ENST00000316577.12
|
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr12_+_56521798 | 0.46 |
ENST00000262031.10
|
RBMS2
|
RNA binding motif single stranded interacting protein 2 |
| chr14_+_75522427 | 0.40 |
ENST00000286639.8
|
BATF
|
basic leucine zipper ATF-like transcription factor |
| chr1_+_202010575 | 0.39 |
ENST00000367283.7
ENST00000367284.10 |
ELF3
|
E74 like ETS transcription factor 3 |
| chr12_-_52796110 | 0.38 |
ENST00000417996.2
|
KRT3
|
keratin 3 |
| chr22_-_17258235 | 0.36 |
ENST00000649310.1
ENST00000649746.1 |
ADA2
|
adenosine deaminase 2 |
| chr7_+_74453790 | 0.35 |
ENST00000265755.7
ENST00000424337.7 ENST00000455841.6 |
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr12_-_89656093 | 0.31 |
ENST00000359142.7
|
ATP2B1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr11_-_119346695 | 0.30 |
ENST00000619721.6
|
MFRP
|
membrane frizzled-related protein |
| chr18_-_49491586 | 0.30 |
ENST00000584895.5
ENST00000580210.5 ENST00000579408.5 |
RPL17-C18orf32
RPL17
|
RPL17-C18orf32 readthrough ribosomal protein L17 |
| chr12_-_89656051 | 0.30 |
ENST00000261173.6
|
ATP2B1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr10_-_48274567 | 0.30 |
ENST00000636244.1
ENST00000374201.8 |
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr9_-_92404559 | 0.29 |
ENST00000262551.8
ENST00000375561.10 |
OGN
|
osteoglycin |
| chr3_+_159069252 | 0.27 |
ENST00000640015.1
ENST00000476809.7 ENST00000485419.7 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr16_-_11976611 | 0.27 |
ENST00000538896.5
ENST00000673243.1 |
NPIPB2
|
nuclear pore complex interacting protein family member B2 |
| chr12_+_6840800 | 0.25 |
ENST00000541978.5
ENST00000229264.8 ENST00000435982.6 |
GNB3
|
G protein subunit beta 3 |
| chr14_+_75522531 | 0.23 |
ENST00000555504.1
|
BATF
|
basic leucine zipper ATF-like transcription factor |
| chr10_+_102226293 | 0.19 |
ENST00000370005.4
|
ELOVL3
|
ELOVL fatty acid elongase 3 |
| chr6_+_35342614 | 0.19 |
ENST00000337400.6
ENST00000311565.4 |
PPARD
|
peroxisome proliferator activated receptor delta |
| chr22_-_29838227 | 0.17 |
ENST00000307790.8
ENST00000397771.6 ENST00000542393.5 |
ASCC2
|
activating signal cointegrator 1 complex subunit 2 |
| chr11_+_61755372 | 0.16 |
ENST00000265460.9
|
MYRF
|
myelin regulatory factor |
| chr16_-_67980483 | 0.14 |
ENST00000268793.6
ENST00000672962.1 |
DPEP3
|
dipeptidase 3 |
| chr7_-_123199960 | 0.11 |
ENST00000194130.7
|
SLC13A1
|
solute carrier family 13 member 1 |
| chr15_+_41774539 | 0.11 |
ENST00000514566.5
|
MAPKBP1
|
mitogen-activated protein kinase binding protein 1 |
| chr17_-_4739866 | 0.10 |
ENST00000574412.6
ENST00000293778.12 |
CXCL16
|
C-X-C motif chemokine ligand 16 |
| chr12_-_118359639 | 0.09 |
ENST00000541786.5
ENST00000419821.6 ENST00000541878.5 |
TAOK3
|
TAO kinase 3 |
| chr6_-_41705813 | 0.09 |
ENST00000419574.6
ENST00000445214.2 |
TFEB
|
transcription factor EB |
| chr22_-_38700655 | 0.09 |
ENST00000216039.9
|
JOSD1
|
Josephin domain containing 1 |
| chr7_+_30284574 | 0.09 |
ENST00000323037.5
|
ZNRF2
|
zinc and ring finger 2 |
| chr8_+_48008409 | 0.07 |
ENST00000523432.5
ENST00000521346.5 ENST00000523111.7 ENST00000517630.5 |
UBE2V2
|
ubiquitin conjugating enzyme E2 V2 |
| chr6_+_46693835 | 0.07 |
ENST00000450697.1
|
TDRD6
|
tudor domain containing 6 |
| chr13_-_35855758 | 0.07 |
ENST00000615680.4
|
DCLK1
|
doublecortin like kinase 1 |
| chr7_-_128775793 | 0.06 |
ENST00000249389.3
|
OPN1SW
|
opsin 1, short wave sensitive |
| chr13_-_35855627 | 0.05 |
ENST00000379893.5
|
DCLK1
|
doublecortin like kinase 1 |
| chr4_-_67754335 | 0.04 |
ENST00000420975.2
ENST00000226413.5 |
GNRHR
|
gonadotropin releasing hormone receptor |
| chr2_+_120013111 | 0.04 |
ENST00000331393.8
ENST00000443124.5 |
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr11_-_5227063 | 0.03 |
ENST00000335295.4
ENST00000485743.1 ENST00000647020.1 |
HBB
|
hemoglobin subunit beta |
| chr3_+_154121366 | 0.03 |
ENST00000465093.6
ENST00000496710.5 ENST00000465817.1 |
ARHGEF26
|
Rho guanine nucleotide exchange factor 26 |
| chr9_-_128724088 | 0.03 |
ENST00000406904.2
ENST00000452105.5 ENST00000372667.9 ENST00000372663.9 |
ZDHHC12
|
zinc finger DHHC-type palmitoyltransferase 12 |
| chr2_+_3575303 | 0.01 |
ENST00000646909.1
ENST00000647131.1 ENST00000462576.5 ENST00000403564.5 |
RPS7
|
ribosomal protein S7 |
| chr22_-_30471986 | 0.01 |
ENST00000401751.5
ENST00000402286.5 ENST00000403066.5 ENST00000215812.9 |
SEC14L3
|
SEC14 like lipid binding 3 |
| chr19_+_47821907 | 0.01 |
ENST00000539067.5
ENST00000221996.12 ENST00000613299.1 |
CRX
|
cone-rod homeobox |
| chr3_+_98497681 | 0.00 |
ENST00000427338.3
|
OR5K2
|
olfactory receptor family 5 subfamily K member 2 |
| chr13_+_73058993 | 0.00 |
ENST00000377687.6
|
KLF5
|
Kruppel like factor 5 |
| chrM_+_9207 | 0.00 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chrX_-_21658324 | 0.00 |
ENST00000379499.3
|
KLHL34
|
kelch like family member 34 |
| chr8_-_132111159 | 0.00 |
ENST00000673615.1
ENST00000434736.6 |
HHLA1
|
HERV-H LTR-associating 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.3 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.1 | 0.8 | GO:1903588 | regulation of neural retina development(GO:0061074) negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.1 | 0.5 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 1.3 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 2.1 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.1 | 0.6 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.1 | 0.4 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.8 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.1 | 0.4 | GO:0046103 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.6 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.0 | 0.4 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.8 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.3 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.2 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.9 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.2 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.1 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.0 | GO:0030185 | nitric oxide transport(GO:0030185) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.8 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 1.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.4 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.8 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 2.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.8 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 1.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |