Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
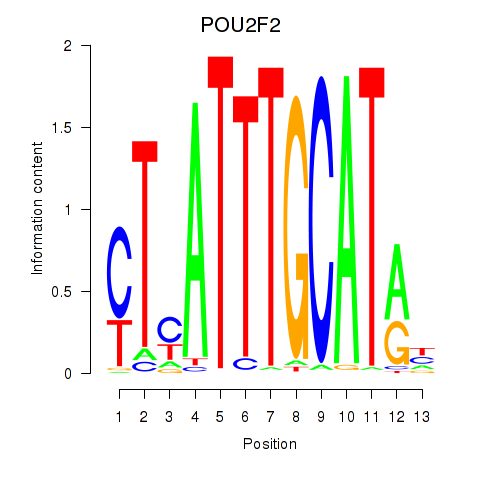
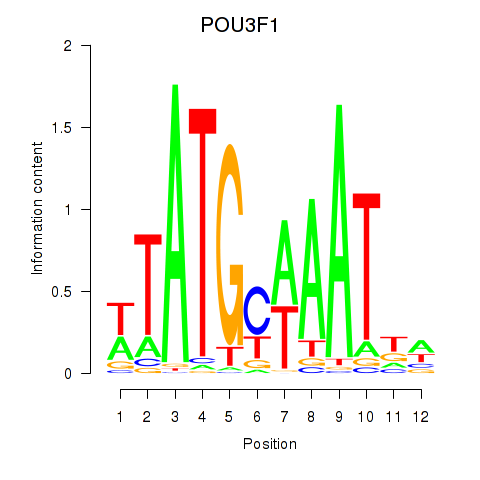
Results for POU2F2_POU3F1
Z-value: 0.68
Transcription factors associated with POU2F2_POU3F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU2F2
|
ENSG00000028277.21 | POU2F2 |
|
POU3F1
|
ENSG00000185668.8 | POU3F1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU2F2 | hg38_v1_chr19_-_42132465_42132480 | 0.56 | 3.5e-03 | Click! |
| POU3F1 | hg38_v1_chr1_-_38046785_38046802 | 0.23 | 2.7e-01 | Click! |
Activity profile of POU2F2_POU3F1 motif
Sorted Z-values of POU2F2_POU3F1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of POU2F2_POU3F1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_67166019 | 1.89 |
ENST00000537194.6
|
SMAD3
|
SMAD family member 3 |
| chr6_+_37170133 | 1.89 |
ENST00000373509.6
|
PIM1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr6_-_142945028 | 1.32 |
ENST00000012134.7
|
HIVEP2
|
HIVEP zinc finger 2 |
| chr6_-_142945160 | 1.31 |
ENST00000367603.8
|
HIVEP2
|
HIVEP zinc finger 2 |
| chr6_-_10415043 | 1.31 |
ENST00000379613.10
|
TFAP2A
|
transcription factor AP-2 alpha |
| chr6_+_31620701 | 1.22 |
ENST00000376033.3
ENST00000376007.8 |
PRRC2A
|
proline rich coiled-coil 2A |
| chr7_+_18496162 | 1.15 |
ENST00000406072.5
|
HDAC9
|
histone deacetylase 9 |
| chr2_+_151357583 | 1.07 |
ENST00000243347.5
|
TNFAIP6
|
TNF alpha induced protein 6 |
| chr6_-_81752671 | 1.00 |
ENST00000320172.11
ENST00000369754.7 ENST00000369756.3 |
TENT5A
|
terminal nucleotidyltransferase 5A |
| chr14_+_64540734 | 0.97 |
ENST00000247207.7
|
HSPA2
|
heat shock protein family A (Hsp70) member 2 |
| chr2_-_224947030 | 0.96 |
ENST00000409592.7
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr2_+_102355750 | 0.86 |
ENST00000233957.7
|
IL18R1
|
interleukin 18 receptor 1 |
| chr1_-_149842736 | 0.84 |
ENST00000369159.2
|
H2AC18
|
H2A clustered histone 18 |
| chr1_+_149851053 | 0.83 |
ENST00000607355.2
|
H2AC19
|
H2A clustered histone 19 |
| chr17_+_20156045 | 0.79 |
ENST00000679801.1
ENST00000679740.1 ENST00000395522.6 ENST00000395525.7 |
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr14_-_106235582 | 0.78 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21 |
| chr1_-_173050931 | 0.78 |
ENST00000404377.5
|
TNFSF18
|
TNF superfamily member 18 |
| chr1_+_67685170 | 0.78 |
ENST00000370985.4
ENST00000370986.9 ENST00000650283.1 ENST00000648742.1 |
GADD45A
|
growth arrest and DNA damage inducible alpha |
| chr14_-_106154113 | 0.76 |
ENST00000390603.2
|
IGHV3-15
|
immunoglobulin heavy variable 3-15 |
| chr1_-_7940825 | 0.75 |
ENST00000377507.8
|
TNFRSF9
|
TNF receptor superfamily member 9 |
| chr20_+_34704336 | 0.71 |
ENST00000374809.6
ENST00000374810.8 ENST00000451665.5 |
TP53INP2
|
tumor protein p53 inducible nuclear protein 2 |
| chr19_-_43619591 | 0.70 |
ENST00000598676.1
ENST00000300811.8 |
ZNF428
|
zinc finger protein 428 |
| chr1_+_167329044 | 0.70 |
ENST00000367862.9
|
POU2F1
|
POU class 2 homeobox 1 |
| chr11_-_62601818 | 0.68 |
ENST00000278823.7
|
MTA2
|
metastasis associated 1 family member 2 |
| chr17_-_7929793 | 0.63 |
ENST00000303790.3
|
KCNAB3
|
potassium voltage-gated channel subfamily A regulatory beta subunit 3 |
| chr10_+_24239181 | 0.60 |
ENST00000438429.5
|
KIAA1217
|
KIAA1217 |
| chr7_+_143222037 | 0.60 |
ENST00000408947.4
|
TAS2R40
|
taste 2 receptor member 40 |
| chr17_+_20155989 | 0.59 |
ENST00000395530.6
ENST00000581399.6 ENST00000679819.1 |
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr20_-_62407274 | 0.58 |
ENST00000279101.9
|
CABLES2
|
Cdk5 and Abl enzyme substrate 2 |
| chr1_+_67685342 | 0.58 |
ENST00000617962.2
|
GADD45A
|
growth arrest and DNA damage inducible alpha |
| chr16_+_33827140 | 0.57 |
ENST00000562905.2
|
IGHV3OR16-13
|
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
| chr2_-_89297785 | 0.57 |
ENST00000465170.1
|
IGKV1-37
|
immunoglobulin kappa variable 1-37 (non-functional) |
| chr18_+_80109236 | 0.55 |
ENST00000262198.9
ENST00000560752.5 |
ADNP2
|
ADNP homeobox 2 |
| chr11_-_133845495 | 0.54 |
ENST00000299140.8
ENST00000532889.1 |
SPATA19
|
spermatogenesis associated 19 |
| chr14_-_106557465 | 0.54 |
ENST00000390625.3
|
IGHV3-49
|
immunoglobulin heavy variable 3-49 |
| chr12_+_112906777 | 0.54 |
ENST00000452357.7
ENST00000445409.7 |
OAS1
|
2'-5'-oligoadenylate synthetase 1 |
| chr6_+_27147094 | 0.54 |
ENST00000377459.3
|
H2AC12
|
H2A clustered histone 12 |
| chr12_+_112906949 | 0.53 |
ENST00000679971.1
ENST00000675868.2 ENST00000550883.2 ENST00000553152.2 ENST00000202917.10 ENST00000679467.1 ENST00000680659.1 ENST00000540589.3 ENST00000552526.2 ENST00000681228.1 ENST00000680934.1 ENST00000681700.1 ENST00000679987.1 |
OAS1
|
2'-5'-oligoadenylate synthetase 1 |
| chr2_+_89884740 | 0.53 |
ENST00000509129.1
|
IGKV1D-37
|
immunoglobulin kappa variable 1D-37 (non-functional) |
| chr15_-_55249029 | 0.53 |
ENST00000566877.5
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr19_+_39412650 | 0.52 |
ENST00000425673.6
|
PLEKHG2
|
pleckstrin homology and RhoGEF domain containing G2 |
| chr8_-_133102874 | 0.52 |
ENST00000395352.7
|
SLA
|
Src like adaptor |
| chr17_+_40015428 | 0.52 |
ENST00000394149.8
ENST00000225474.6 ENST00000331769.6 ENST00000394148.7 ENST00000577675.1 |
CSF3
|
colony stimulating factor 3 |
| chr12_+_112907006 | 0.52 |
ENST00000680455.1
ENST00000551241.6 ENST00000550689.2 ENST00000679841.1 ENST00000679494.1 ENST00000553185.2 |
OAS1
|
2'-5'-oligoadenylate synthetase 1 |
| chr6_+_29100609 | 0.51 |
ENST00000377171.3
|
OR2J1
|
olfactory receptor family 2 subfamily J member 1 |
| chr6_-_10414985 | 0.51 |
ENST00000466073.5
ENST00000498450.3 |
TFAP2A
|
transcription factor AP-2 alpha |
| chr6_-_26123910 | 0.51 |
ENST00000314332.5
ENST00000396984.1 |
H2BC4
|
H2B clustered histone 4 |
| chr3_-_71583713 | 0.50 |
ENST00000649528.3
ENST00000471386.3 ENST00000493089.7 |
FOXP1
|
forkhead box P1 |
| chr7_+_18495723 | 0.49 |
ENST00000681950.1
ENST00000622668.4 ENST00000405010.7 ENST00000406451.8 ENST00000441542.7 ENST00000428307.6 ENST00000681273.1 |
HDAC9
|
histone deacetylase 9 |
| chr2_-_206218024 | 0.48 |
ENST00000407325.6
ENST00000612892.4 ENST00000411719.1 |
GPR1
|
G protein-coupled receptor 1 |
| chr2_+_89913982 | 0.48 |
ENST00000390265.2
|
IGKV1D-33
|
immunoglobulin kappa variable 1D-33 |
| chr1_-_2526585 | 0.47 |
ENST00000378466.9
ENST00000435556.8 |
PANK4
|
pantothenate kinase 4 (inactive) |
| chr6_-_33711717 | 0.46 |
ENST00000374214.3
|
UQCC2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr6_-_138499487 | 0.44 |
ENST00000343505.9
|
NHSL1
|
NHS like 1 |
| chr2_+_219279330 | 0.42 |
ENST00000425450.5
ENST00000392086.8 ENST00000421532.5 ENST00000336576.10 |
DNAJB2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr6_-_27132750 | 0.42 |
ENST00000607124.1
ENST00000339812.3 |
H2BC11
|
H2B clustered histone 11 |
| chr2_+_85429448 | 0.41 |
ENST00000651736.1
|
SH2D6
|
SH2 domain containing 6 |
| chr2_-_89268506 | 0.40 |
ENST00000473726.1
|
IGKV1-33
|
immunoglobulin kappa variable 1-33 |
| chr6_+_26251607 | 0.40 |
ENST00000619466.2
|
H2BC9
|
H2B clustered histone 9 |
| chr3_-_71130557 | 0.40 |
ENST00000497355.7
|
FOXP1
|
forkhead box P1 |
| chr3_-_108058361 | 0.40 |
ENST00000398258.7
|
CD47
|
CD47 molecule |
| chr11_-_3642273 | 0.39 |
ENST00000359918.8
|
ART5
|
ADP-ribosyltransferase 5 |
| chr19_-_45584810 | 0.39 |
ENST00000323060.3
|
OPA3
|
outer mitochondrial membrane lipid metabolism regulator OPA3 |
| chr6_+_10585748 | 0.39 |
ENST00000265012.5
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2 (I blood group) |
| chr11_+_69641146 | 0.38 |
ENST00000227507.3
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr19_+_11355386 | 0.38 |
ENST00000251473.9
ENST00000591329.5 ENST00000586380.5 |
PLPPR2
|
phospholipid phosphatase related 2 |
| chr16_+_33009175 | 0.37 |
ENST00000565407.2
|
IGHV3OR16-8
|
immunoglobulin heavy variable 3/OR16-8 (non-functional) |
| chr6_+_27893411 | 0.37 |
ENST00000616182.2
|
H2BC17
|
H2B clustered histone 17 |
| chr7_-_100586119 | 0.37 |
ENST00000310300.11
|
LRCH4
|
leucine rich repeats and calponin homology domain containing 4 |
| chr1_+_149886906 | 0.37 |
ENST00000331380.4
|
H2AC20
|
H2A clustered histone 20 |
| chr3_-_71130892 | 0.36 |
ENST00000491238.7
ENST00000674446.1 |
FOXP1
|
forkhead box P1 |
| chr6_-_27893175 | 0.36 |
ENST00000359611.4
|
H2AC17
|
H2A clustered histone 17 |
| chr8_-_26045360 | 0.36 |
ENST00000520164.6
|
EBF2
|
EBF transcription factor 2 |
| chr19_+_11355491 | 0.36 |
ENST00000591608.1
|
PLPPR2
|
phospholipid phosphatase related 2 |
| chr19_-_1132208 | 0.36 |
ENST00000438103.6
|
SBNO2
|
strawberry notch homolog 2 |
| chr6_-_53148822 | 0.36 |
ENST00000259803.8
|
GCM1
|
glial cells missing transcription factor 1 |
| chr3_-_123884290 | 0.36 |
ENST00000346322.9
ENST00000360772.7 ENST00000360304.8 |
MYLK
|
myosin light chain kinase |
| chr6_-_33711684 | 0.36 |
ENST00000374231.8
ENST00000607484.6 |
UQCC2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr3_-_71130963 | 0.36 |
ENST00000649695.2
|
FOXP1
|
forkhead box P1 |
| chr6_-_26216673 | 0.35 |
ENST00000541790.3
|
H2BC8
|
H2B clustered histone 8 |
| chr1_-_10964201 | 0.34 |
ENST00000418570.6
|
C1orf127
|
chromosome 1 open reading frame 127 |
| chr2_+_90154073 | 0.34 |
ENST00000611391.1
|
IGKV1D-13
|
immunoglobulin kappa variable 1D-13 |
| chr19_-_17405554 | 0.34 |
ENST00000252593.7
|
BST2
|
bone marrow stromal cell antigen 2 |
| chr9_+_89605004 | 0.34 |
ENST00000252506.11
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA damage inducible gamma |
| chr14_+_103107516 | 0.34 |
ENST00000560304.1
|
EXOC3L4
|
exocyst complex component 3 like 4 |
| chr19_-_14884761 | 0.34 |
ENST00000642123.1
|
OR7A17
|
olfactory receptor family 7 subfamily A member 17 |
| chr2_+_54558348 | 0.34 |
ENST00000333896.5
|
SPTBN1
|
spectrin beta, non-erythrocytic 1 |
| chr16_+_56935371 | 0.31 |
ENST00000568358.1
|
HERPUD1
|
homocysteine inducible ER protein with ubiquitin like domain 1 |
| chr2_+_173354820 | 0.31 |
ENST00000347703.7
ENST00000410101.7 ENST00000410019.3 ENST00000306721.8 |
CDCA7
|
cell division cycle associated 7 |
| chr2_-_171894227 | 0.31 |
ENST00000422440.7
|
SLC25A12
|
solute carrier family 25 member 12 |
| chr19_-_893172 | 0.31 |
ENST00000325464.6
ENST00000312090.10 |
MED16
|
mediator complex subunit 16 |
| chr1_+_59310071 | 0.31 |
ENST00000371212.5
|
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr16_-_33845229 | 0.30 |
ENST00000569103.2
|
IGHV3OR16-17
|
immunoglobulin heavy variable 3/OR16-17 (non-functional) |
| chr14_-_106211453 | 0.30 |
ENST00000390606.3
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr22_+_22811737 | 0.30 |
ENST00000390315.3
|
IGLV3-10
|
immunoglobulin lambda variable 3-10 |
| chr19_-_893200 | 0.29 |
ENST00000269814.8
ENST00000395808.7 |
MED16
|
mediator complex subunit 16 |
| chr21_-_30829755 | 0.29 |
ENST00000621162.1
|
KRTAP7-1
|
keratin associated protein 7-1 |
| chr17_+_3207539 | 0.29 |
ENST00000641322.1
ENST00000641732.2 |
OR1A1
|
olfactory receptor family 1 subfamily A member 1 |
| chr14_-_106791226 | 0.29 |
ENST00000433072.2
|
IGHV3-72
|
immunoglobulin heavy variable 3-72 |
| chr1_-_183653307 | 0.28 |
ENST00000308641.6
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme catalytic polypeptide like 4 |
| chr22_+_22030934 | 0.28 |
ENST00000390282.2
|
IGLV4-69
|
immunoglobulin lambda variable 4-69 |
| chr19_-_18397596 | 0.27 |
ENST00000595840.1
ENST00000339007.4 |
LRRC25
|
leucine rich repeat containing 25 |
| chr8_-_33513077 | 0.27 |
ENST00000360742.9
ENST00000523305.1 ENST00000431156.7 ENST00000613904.1 |
TTI2
|
TELO2 interacting protein 2 |
| chr5_-_67196791 | 0.27 |
ENST00000256447.5
|
CD180
|
CD180 molecule |
| chr8_-_133102477 | 0.26 |
ENST00000522119.5
ENST00000523610.5 ENST00000338087.10 ENST00000521302.5 ENST00000519558.5 ENST00000519747.5 ENST00000517648.5 |
SLA
|
Src like adaptor |
| chr1_-_201115372 | 0.26 |
ENST00000458416.2
|
ASCL5
|
achaete-scute family bHLH transcription factor 5 |
| chr17_-_37745018 | 0.26 |
ENST00000613727.4
ENST00000614313.4 ENST00000617811.5 ENST00000621123.4 |
HNF1B
|
HNF1 homeobox B |
| chr6_+_155216637 | 0.26 |
ENST00000275246.11
|
TIAM2
|
TIAM Rac1 associated GEF 2 |
| chr17_-_75131729 | 0.26 |
ENST00000245552.7
ENST00000582170.1 |
NT5C
|
5', 3'-nucleotidase, cytosolic |
| chr17_+_7650916 | 0.26 |
ENST00000250111.9
|
ATP1B2
|
ATPase Na+/K+ transporting subunit beta 2 |
| chr5_+_126423122 | 0.26 |
ENST00000515200.5
|
GRAMD2B
|
GRAM domain containing 2B |
| chr15_-_58065703 | 0.25 |
ENST00000249750.9
|
ALDH1A2
|
aldehyde dehydrogenase 1 family member A2 |
| chr14_-_24146314 | 0.25 |
ENST00000559056.5
|
PSME2
|
proteasome activator subunit 2 |
| chr7_+_128672275 | 0.25 |
ENST00000684498.1
ENST00000641605.1 ENST00000682356.1 ENST00000477515.3 |
FAM71F2
|
family with sequence similarity 71 member F2 |
| chr8_-_25458389 | 0.25 |
ENST00000221200.9
|
KCTD9
|
potassium channel tetramerization domain containing 9 |
| chr20_+_836052 | 0.25 |
ENST00000246100.3
|
FAM110A
|
family with sequence similarity 110 member A |
| chr5_+_126423176 | 0.25 |
ENST00000542322.5
ENST00000544396.5 |
GRAMD2B
|
GRAM domain containing 2B |
| chr14_-_106130061 | 0.25 |
ENST00000390602.3
|
IGHV3-13
|
immunoglobulin heavy variable 3-13 |
| chr18_-_77127935 | 0.25 |
ENST00000581878.5
|
MBP
|
myelin basic protein |
| chr5_-_159099909 | 0.25 |
ENST00000313708.11
|
EBF1
|
EBF transcription factor 1 |
| chr5_+_76819022 | 0.25 |
ENST00000296677.5
|
F2RL1
|
F2R like trypsin receptor 1 |
| chr5_+_126423363 | 0.25 |
ENST00000285689.8
|
GRAMD2B
|
GRAM domain containing 2B |
| chr19_+_42284020 | 0.25 |
ENST00000160740.7
|
CIC
|
capicua transcriptional repressor |
| chr4_-_47981535 | 0.25 |
ENST00000402813.9
|
CNGA1
|
cyclic nucleotide gated channel subunit alpha 1 |
| chr21_-_34526850 | 0.25 |
ENST00000481448.5
ENST00000381132.6 |
RCAN1
|
regulator of calcineurin 1 |
| chrX_+_38801451 | 0.25 |
ENST00000378474.3
ENST00000336949.7 |
MID1IP1
|
MID1 interacting protein 1 |
| chr15_-_58065734 | 0.24 |
ENST00000347587.7
|
ALDH1A2
|
aldehyde dehydrogenase 1 family member A2 |
| chr10_+_96000091 | 0.24 |
ENST00000424464.5
ENST00000410012.6 ENST00000344386.3 |
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chr5_+_80960694 | 0.24 |
ENST00000638442.1
|
RASGRF2
|
Ras protein specific guanine nucleotide releasing factor 2 |
| chr1_+_166839425 | 0.24 |
ENST00000449930.5
ENST00000367876.9 |
POGK
|
pogo transposable element derived with KRAB domain |
| chr19_+_19033575 | 0.24 |
ENST00000392335.6
ENST00000537263.5 ENST00000540707.5 ENST00000535612.6 ENST00000541725.5 ENST00000269932.10 ENST00000546344.5 ENST00000540792.5 ENST00000536098.5 ENST00000541898.5 |
ARMC6
|
armadillo repeat containing 6 |
| chr12_-_10998304 | 0.23 |
ENST00000538986.2
|
TAS2R20
|
taste 2 receptor member 20 |
| chr10_+_104353820 | 0.23 |
ENST00000369704.8
|
CFAP58
|
cilia and flagella associated protein 58 |
| chr12_-_16606795 | 0.23 |
ENST00000447609.5
|
LMO3
|
LIM domain only 3 |
| chr11_+_64318091 | 0.23 |
ENST00000265462.9
ENST00000352435.8 ENST00000347941.4 |
PRDX5
|
peroxiredoxin 5 |
| chr19_-_19170248 | 0.23 |
ENST00000410050.5
ENST00000424583.7 ENST00000409224.5 ENST00000409447.2 |
MEF2B
|
myocyte enhancer factor 2B |
| chr11_+_27055215 | 0.23 |
ENST00000525090.1
|
BBOX1
|
gamma-butyrobetaine hydroxylase 1 |
| chr12_-_7092422 | 0.23 |
ENST00000543835.5
ENST00000647956.2 ENST00000535233.6 |
C1R
|
complement C1r |
| chr19_-_45584769 | 0.22 |
ENST00000263275.5
|
OPA3
|
outer mitochondrial membrane lipid metabolism regulator OPA3 |
| chr17_-_48817217 | 0.22 |
ENST00000393382.8
|
TTLL6
|
tubulin tyrosine ligase like 6 |
| chrX_+_38801417 | 0.22 |
ENST00000614558.2
ENST00000457894.5 |
MID1IP1
|
MID1 interacting protein 1 |
| chr3_+_63443306 | 0.22 |
ENST00000472899.5
ENST00000479198.5 ENST00000460711.5 ENST00000465156.1 |
SYNPR
|
synaptoporin |
| chr18_-_63644250 | 0.22 |
ENST00000341074.10
ENST00000436264.1 |
SERPINB4
|
serpin family B member 4 |
| chr5_-_36301883 | 0.22 |
ENST00000502994.5
ENST00000515759.5 ENST00000296604.8 |
RANBP3L
|
RAN binding protein 3 like |
| chr19_+_40799155 | 0.22 |
ENST00000303961.9
|
EGLN2
|
egl-9 family hypoxia inducible factor 2 |
| chr4_-_109302643 | 0.21 |
ENST00000399126.1
ENST00000505591.1 ENST00000399132.6 |
COL25A1
|
collagen type XXV alpha 1 chain |
| chr20_+_56629296 | 0.21 |
ENST00000201031.3
|
TFAP2C
|
transcription factor AP-2 gamma |
| chr15_-_58065870 | 0.21 |
ENST00000537372.5
|
ALDH1A2
|
aldehyde dehydrogenase 1 family member A2 |
| chr3_+_63443076 | 0.21 |
ENST00000295894.9
|
SYNPR
|
synaptoporin |
| chr6_+_29301701 | 0.21 |
ENST00000641895.1
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1 |
| chr2_+_90172802 | 0.20 |
ENST00000390277.3
|
IGKV3D-11
|
immunoglobulin kappa variable 3D-11 |
| chr19_+_35358460 | 0.20 |
ENST00000327809.5
|
FFAR3
|
free fatty acid receptor 3 |
| chr15_+_80441229 | 0.20 |
ENST00000533983.5
ENST00000527771.5 ENST00000525103.1 |
ARNT2
|
aryl hydrocarbon receptor nuclear translocator 2 |
| chr21_-_34888683 | 0.20 |
ENST00000344691.8
ENST00000358356.9 |
RUNX1
|
RUNX family transcription factor 1 |
| chr10_+_122163426 | 0.20 |
ENST00000360561.7
|
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chr3_-_71583683 | 0.20 |
ENST00000649631.1
ENST00000648718.1 |
FOXP1
|
forkhead box P1 |
| chr17_-_3063607 | 0.20 |
ENST00000575751.1
|
OR1D5
|
olfactory receptor family 1 subfamily D member 5 |
| chr1_-_196608359 | 0.19 |
ENST00000609185.5
ENST00000451324.6 ENST00000367433.9 ENST00000294725.14 |
KCNT2
|
potassium sodium-activated channel subfamily T member 2 |
| chr10_+_122163590 | 0.19 |
ENST00000368999.5
|
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chr2_+_113406368 | 0.19 |
ENST00000453673.3
|
IGKV1OR2-108
|
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chr10_+_122163672 | 0.19 |
ENST00000369004.7
ENST00000260733.7 |
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chr19_-_46788586 | 0.19 |
ENST00000542575.6
|
SLC1A5
|
solute carrier family 1 member 5 |
| chr11_+_27041313 | 0.19 |
ENST00000528583.5
|
BBOX1
|
gamma-butyrobetaine hydroxylase 1 |
| chr6_+_26183750 | 0.19 |
ENST00000614097.3
|
H2BC6
|
H2B clustered histone 6 |
| chr3_-_71583592 | 0.19 |
ENST00000650156.1
ENST00000649596.1 |
FOXP1
|
forkhead box P1 |
| chr2_-_89117844 | 0.19 |
ENST00000490686.1
|
IGKV1-17
|
immunoglobulin kappa variable 1-17 |
| chr2_-_88966767 | 0.19 |
ENST00000464162.1
|
IGKV1-6
|
immunoglobulin kappa variable 1-6 |
| chr15_-_66356672 | 0.19 |
ENST00000261881.9
|
TIPIN
|
TIMELESS interacting protein |
| chr7_+_123601815 | 0.19 |
ENST00000451215.6
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr1_-_165445088 | 0.19 |
ENST00000359842.10
|
RXRG
|
retinoid X receptor gamma |
| chr12_+_107318395 | 0.18 |
ENST00000420571.6
ENST00000280758.10 |
BTBD11
|
BTB domain containing 11 |
| chr1_+_156150008 | 0.18 |
ENST00000355014.6
|
SEMA4A
|
semaphorin 4A |
| chr6_-_127900958 | 0.18 |
ENST00000434358.3
ENST00000630369.2 ENST00000368248.4 |
THEMIS
|
thymocyte selection associated |
| chr17_-_75131683 | 0.18 |
ENST00000578407.5
|
NT5C
|
5', 3'-nucleotidase, cytosolic |
| chr1_+_207496268 | 0.18 |
ENST00000529814.1
|
CR1
|
complement C3b/C4b receptor 1 (Knops blood group) |
| chr1_+_27773189 | 0.18 |
ENST00000373943.9
ENST00000440806.2 |
STX12
|
syntaxin 12 |
| chr10_+_110005804 | 0.18 |
ENST00000360162.7
|
ADD3
|
adducin 3 |
| chr11_-_18236795 | 0.18 |
ENST00000278222.7
|
SAA4
|
serum amyloid A4, constitutive |
| chr1_+_32200587 | 0.18 |
ENST00000373602.10
ENST00000421922.6 ENST00000681230.1 |
CCDC28B
|
coiled-coil domain containing 28B |
| chr7_+_123601836 | 0.18 |
ENST00000434204.5
|
ASB15
|
ankyrin repeat and SOCS box containing 15 |
| chr7_-_113087720 | 0.18 |
ENST00000297146.7
|
GPR85
|
G protein-coupled receptor 85 |
| chr2_-_157325808 | 0.18 |
ENST00000410096.6
ENST00000420719.6 ENST00000409216.5 ENST00000419116.2 |
ERMN
|
ermin |
| chr6_-_43687774 | 0.18 |
ENST00000372133.8
ENST00000372116.5 ENST00000427312.1 |
MRPS18A
|
mitochondrial ribosomal protein S18A |
| chr6_+_41042462 | 0.17 |
ENST00000373161.6
|
TSPO2
|
translocator protein 2 |
| chr11_-_34357994 | 0.17 |
ENST00000435224.3
|
ABTB2
|
ankyrin repeat and BTB domain containing 2 |
| chr2_-_89100352 | 0.17 |
ENST00000479981.1
|
IGKV1-16
|
immunoglobulin kappa variable 1-16 |
| chr17_-_10804092 | 0.17 |
ENST00000581851.1
|
TMEM238L
|
transmembrane protein 238 like |
| chr14_-_50830479 | 0.17 |
ENST00000382043.8
|
NIN
|
ninein |
| chr11_+_27040725 | 0.17 |
ENST00000529202.5
ENST00000263182.8 |
BBOX1
|
gamma-butyrobetaine hydroxylase 1 |
| chr15_-_55917080 | 0.17 |
ENST00000506154.1
|
NEDD4
|
NEDD4 E3 ubiquitin protein ligase |
| chr11_-_6030758 | 0.16 |
ENST00000641900.1
|
OR56A1
|
olfactory receptor family 56 subfamily A member 1 |
| chr10_-_62236112 | 0.16 |
ENST00000315289.6
|
RTKN2
|
rhotekin 2 |
| chr2_+_90234809 | 0.16 |
ENST00000443397.5
|
IGKV3D-7
|
immunoglobulin kappa variable 3D-7 |
| chr22_-_23754376 | 0.16 |
ENST00000398465.3
ENST00000248948.4 |
VPREB3
|
V-set pre-B cell surrogate light chain 3 |
| chr22_+_22734577 | 0.16 |
ENST00000390310.3
|
IGLV2-18
|
immunoglobulin lambda variable 2-18 |
| chr12_-_10909562 | 0.15 |
ENST00000390677.2
|
TAS2R13
|
taste 2 receptor member 13 |
| chr2_+_200308943 | 0.15 |
ENST00000619961.4
|
SPATS2L
|
spermatogenesis associated serine rich 2 like |
| chr16_-_8975320 | 0.15 |
ENST00000673704.1
|
USP7
|
ubiquitin specific peptidase 7 |
| chr19_-_3971051 | 0.15 |
ENST00000545797.7
ENST00000596311.5 |
DAPK3
|
death associated protein kinase 3 |
| chr3_+_119597874 | 0.15 |
ENST00000488919.5
ENST00000273371.9 ENST00000495992.5 |
PLA1A
|
phospholipase A1 member A |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.8 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.3 | 1.9 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.2 | 0.8 | GO:2000329 | negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.2 | 1.9 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.1 | 0.4 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 2.0 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 1.6 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.3 | GO:0045957 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) |
| 0.1 | 0.3 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.1 | 0.7 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.1 | 0.9 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) |
| 0.1 | 0.3 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.3 | GO:0044007 | negative regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010768) transmission of virus(GO:0019089) dissemination or transmission of symbiont from host(GO:0044007) dissemination or transmission of organism from other organism involved in symbiotic interaction(GO:0051821) |
| 0.1 | 0.4 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.1 | 0.3 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.1 | 0.3 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 0.1 | 0.5 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.3 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 0.7 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 | 0.8 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.1 | 0.3 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.1 | 0.2 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 0.5 | GO:0035926 | chemokine (C-C motif) ligand 2 secretion(GO:0035926) |
| 0.1 | 0.2 | GO:1990009 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) retinal cell apoptotic process(GO:1990009) |
| 0.1 | 1.5 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.1 | 0.3 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 0.4 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.1 | 1.0 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.4 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 0.6 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.4 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.4 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.2 | GO:0036229 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 0.1 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.3 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.4 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 1.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.5 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:1900222 | positive regulation of transcription via serum response element binding(GO:0010735) negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.0 | 0.1 | GO:0060031 | mediolateral intercalation(GO:0060031) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) |
| 0.0 | 0.3 | GO:0060844 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) blood vessel endothelial cell fate specification(GO:0097101) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:2000317 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.0 | 2.4 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.4 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.1 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.0 | 0.8 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.1 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.1 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.3 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.0 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.2 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.5 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.1 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.0 | 0.5 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.4 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.4 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.3 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.1 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.0 | 0.4 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.1 | GO:0001978 | regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) baroreceptor response to increased systemic arterial blood pressure(GO:0001983) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.0 | 0.6 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.0 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 1.6 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.7 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.0 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.1 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.0 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.7 | GO:0010762 | regulation of fibroblast migration(GO:0010762) |
| 0.0 | 0.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0070166 | enamel mineralization(GO:0070166) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 1.0 | GO:0072687 | CatSper complex(GO:0036128) meiotic spindle(GO:0072687) |
| 0.1 | 0.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 2.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.3 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.4 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 2.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.8 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 1.1 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.3 | 0.9 | GO:0042008 | interleukin-18 receptor activity(GO:0042008) |
| 0.2 | 0.6 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.6 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.3 | GO:0001861 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.1 | 1.6 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.3 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.1 | 1.9 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.4 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.1 | 1.6 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.3 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 1.0 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.4 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.2 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.0 | 2.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.7 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.5 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.0 | 0.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.3 | GO:0050816 | phosphoserine binding(GO:0050815) phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0008893 | guanosine-3',5'-bis(diphosphate) 3'-diphosphatase activity(GO:0008893) diphosphoric monoester hydrolase activity(GO:0016794) |
| 0.0 | 0.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 1.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0052854 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 0.0 | 0.2 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.7 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.2 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 1.9 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 2.3 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.5 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.7 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.0 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.0 | 0.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.0 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.0 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.7 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 1.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |