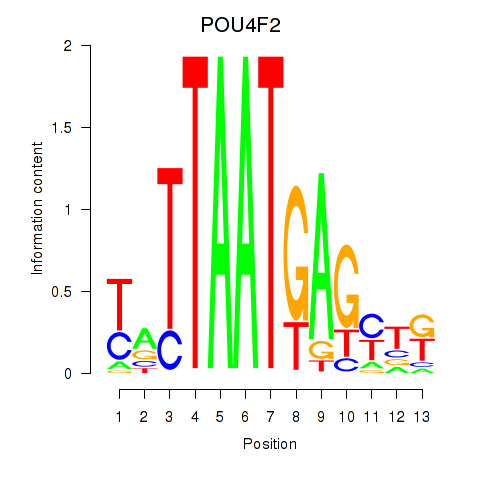
|
chr21_-_30487436
Show fit
|
0.72 |
ENST00000334055.5
|
KRTAP19-2
|
keratin associated protein 19-2
|
|
chr1_-_182672232
Show fit
|
0.58 |
ENST00000508450.5
|
RGS8
|
regulator of G protein signaling 8
|
|
chr19_+_45498439
Show fit
|
0.43 |
ENST00000451287.7
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent 1N (putative)
|
|
chr8_-_18684033
Show fit
|
0.41 |
ENST00000614430.3
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr8_-_18684093
Show fit
|
0.40 |
ENST00000428502.6
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr12_-_52385649
Show fit
|
0.39 |
ENST00000257951.3
|
KRT84
|
keratin 84
|
|
chr1_-_44017296
Show fit
|
0.38 |
ENST00000357730.6
ENST00000360584.6
ENST00000528803.1
|
SLC6A9
|
solute carrier family 6 member 9
|
|
chr8_-_18683932
Show fit
|
0.36 |
ENST00000615573.4
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr3_-_79767987
Show fit
|
0.27 |
ENST00000464233.6
|
ROBO1
|
roundabout guidance receptor 1
|
|
chr9_+_74615582
Show fit
|
0.25 |
ENST00000396204.2
|
RORB
|
RAR related orphan receptor B
|
|
chrX_+_30235894
Show fit
|
0.23 |
ENST00000620842.1
|
MAGEB3
|
MAGE family member B3
|
|
chr6_-_89217339
Show fit
|
0.22 |
ENST00000454853.7
|
GABRR1
|
gamma-aminobutyric acid type A receptor subunit rho1
|
|
chr5_+_90640718
Show fit
|
0.22 |
ENST00000640403.1
|
ADGRV1
|
adhesion G protein-coupled receptor V1
|
|
chr4_+_158521937
Show fit
|
0.21 |
ENST00000343542.9
ENST00000470033.2
|
RXFP1
|
relaxin family peptide receptor 1
|
|
chr2_-_70553440
Show fit
|
0.21 |
ENST00000450929.5
|
TGFA
|
transforming growth factor alpha
|
|
chr12_-_30735014
Show fit
|
0.21 |
ENST00000433722.6
|
CAPRIN2
|
caprin family member 2
|
|
chr6_+_39792298
Show fit
|
0.20 |
ENST00000633794.1
ENST00000274867.9
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr4_+_75556048
Show fit
|
0.20 |
ENST00000616557.1
ENST00000435974.2
ENST00000311623.9
|
ODAPH
|
odontogenesis associated phosphoprotein
|
|
chr10_+_97584314
Show fit
|
0.19 |
ENST00000370647.8
|
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1
|
|
chr18_-_28036585
Show fit
|
0.19 |
ENST00000399380.7
|
CDH2
|
cadherin 2
|
|
chr14_+_21887848
Show fit
|
0.19 |
ENST00000390437.2
|
TRAV12-2
|
T cell receptor alpha variable 12-2
|
|
chr21_-_30492008
Show fit
|
0.18 |
ENST00000334063.6
|
KRTAP19-3
|
keratin associated protein 19-3
|
|
chr11_-_31509569
Show fit
|
0.17 |
ENST00000526776.5
|
IMMP1L
|
inner mitochondrial membrane peptidase subunit 1
|
|
chr17_+_70104848
Show fit
|
0.16 |
ENST00000392670.5
|
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16
|
|
chr6_-_25874212
Show fit
|
0.16 |
ENST00000361703.10
ENST00000397060.8
|
SLC17A3
|
solute carrier family 17 member 3
|
|
chr9_+_122371036
Show fit
|
0.15 |
ENST00000619306.5
ENST00000426608.6
ENST00000223423.8
|
PTGS1
|
prostaglandin-endoperoxide synthase 1
|
|
chr5_-_135578983
Show fit
|
0.15 |
ENST00000512158.6
|
CXCL14
|
C-X-C motif chemokine ligand 14
|
|
chr14_+_31561376
Show fit
|
0.15 |
ENST00000550649.5
ENST00000281081.12
|
NUBPL
|
nucleotide binding protein like
|
|
chr3_-_51875597
Show fit
|
0.15 |
ENST00000446461.2
|
IQCF5
|
IQ motif containing F5
|
|
chr2_-_49973939
Show fit
|
0.13 |
ENST00000630656.1
|
NRXN1
|
neurexin 1
|
|
chr5_-_58999885
Show fit
|
0.13 |
ENST00000317118.12
|
PDE4D
|
phosphodiesterase 4D
|
|
chr4_-_122621011
Show fit
|
0.12 |
ENST00000611104.2
ENST00000648588.1
|
IL21
|
interleukin 21
|
|
chrX_-_100732100
Show fit
|
0.12 |
ENST00000372981.1
ENST00000263033.9
|
SYTL4
|
synaptotagmin like 4
|
|
chr3_-_58627567
Show fit
|
0.12 |
ENST00000649301.1
|
FAM107A
|
family with sequence similarity 107 member A
|
|
chr4_-_168480477
Show fit
|
0.11 |
ENST00000514748.5
ENST00000512371.1
ENST00000505890.5
ENST00000682922.1
ENST00000511577.5
|
DDX60L
|
DExD/H-box 60 like
|
|
chr12_-_7444139
Show fit
|
0.11 |
ENST00000416109.2
ENST00000313599.8
|
CD163L1
|
CD163 molecule like 1
|
|
chr1_+_198638968
Show fit
|
0.11 |
ENST00000348564.11
ENST00000530727.5
ENST00000442510.8
ENST00000645247.1
ENST00000367367.8
ENST00000367364.5
ENST00000413409.6
|
PTPRC
|
protein tyrosine phosphatase receptor type C
|
|
chr17_+_18183052
Show fit
|
0.11 |
ENST00000541285.1
|
ALKBH5
|
alkB homolog 5, RNA demethylase
|
|
chr17_+_18183803
Show fit
|
0.11 |
ENST00000399138.5
|
ALKBH5
|
alkB homolog 5, RNA demethylase
|
|
chr5_+_155013755
Show fit
|
0.11 |
ENST00000435029.6
|
KIF4B
|
kinesin family member 4B
|
|
chr7_-_81770122
Show fit
|
0.10 |
ENST00000423064.7
|
HGF
|
hepatocyte growth factor
|
|
chr11_-_41459592
Show fit
|
0.10 |
ENST00000528697.6
ENST00000530763.5
|
LRRC4C
|
leucine rich repeat containing 4C
|
|
chr7_+_143284930
Show fit
|
0.10 |
ENST00000409244.5
ENST00000409541.5
ENST00000410004.1
ENST00000359333.8
|
TMEM139
|
transmembrane protein 139
|
|
chr8_+_22999535
Show fit
|
0.10 |
ENST00000251822.7
|
RHOBTB2
|
Rho related BTB domain containing 2
|
|
chr4_-_47981535
Show fit
|
0.09 |
ENST00000402813.9
|
CNGA1
|
cyclic nucleotide gated channel subunit alpha 1
|
|
chr12_-_118359639
Show fit
|
0.09 |
ENST00000541786.5
ENST00000419821.6
ENST00000541878.5
|
TAOK3
|
TAO kinase 3
|
|
chr6_+_130018565
Show fit
|
0.09 |
ENST00000361794.7
ENST00000526087.5
ENST00000533560.5
|
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3
|
|
chr16_-_20404731
Show fit
|
0.09 |
ENST00000302451.9
|
PDILT
|
protein disulfide isomerase like, testis expressed
|
|
chr11_-_125680831
Show fit
|
0.09 |
ENST00000315608.7
ENST00000530048.5
ENST00000533904.6
|
ACRV1
|
acrosomal vesicle protein 1
|
|
chr9_+_122370523
Show fit
|
0.09 |
ENST00000643810.1
ENST00000540753.6
|
PTGS1
|
prostaglandin-endoperoxide synthase 1
|
|
chr5_+_135579193
Show fit
|
0.08 |
ENST00000646290.1
|
SLC25A48
|
solute carrier family 25 member 48
|
|
chrX_+_134796758
Show fit
|
0.08 |
ENST00000414371.6
|
PABIR3
|
PABIR family member 3
|
|
chr14_+_19743571
Show fit
|
0.08 |
ENST00000642117.2
|
OR4Q3
|
olfactory receptor family 4 subfamily Q member 3
|
|
chr9_+_122371014
Show fit
|
0.07 |
ENST00000362012.7
|
PTGS1
|
prostaglandin-endoperoxide synthase 1
|
|
chr15_-_37101205
Show fit
|
0.07 |
ENST00000338564.9
ENST00000558313.5
ENST00000340545.9
|
MEIS2
|
Meis homeobox 2
|
|
chr11_-_125680869
Show fit
|
0.07 |
ENST00000527795.1
|
ACRV1
|
acrosomal vesicle protein 1
|
|
chr4_+_158521714
Show fit
|
0.06 |
ENST00000613319.4
ENST00000423548.5
ENST00000448688.6
|
RXFP1
|
relaxin family peptide receptor 1
|
|
chr8_+_49911604
Show fit
|
0.06 |
ENST00000642164.1
ENST00000644093.1
ENST00000643999.1
ENST00000647073.1
ENST00000646880.1
|
SNTG1
|
syntrophin gamma 1
|
|
chr15_+_45252228
Show fit
|
0.06 |
ENST00000560438.5
ENST00000347644.8
|
SLC28A2
|
solute carrier family 28 member 2
|
|
chrX_-_106903331
Show fit
|
0.06 |
ENST00000411805.1
ENST00000276173.5
|
RIPPLY1
|
ripply transcriptional repressor 1
|
|
chr17_-_66229380
Show fit
|
0.06 |
ENST00000205948.11
|
APOH
|
apolipoprotein H
|
|
chr12_-_118359105
Show fit
|
0.05 |
ENST00000541186.5
ENST00000539872.5
|
TAOK3
|
TAO kinase 3
|
|
chr8_-_48921419
Show fit
|
0.05 |
ENST00000020945.4
|
SNAI2
|
snail family transcriptional repressor 2
|
|
chr21_-_30813270
Show fit
|
0.05 |
ENST00000329621.6
|
KRTAP8-1
|
keratin associated protein 8-1
|
|
chr4_+_158521872
Show fit
|
0.05 |
ENST00000307765.10
|
RXFP1
|
relaxin family peptide receptor 1
|
|
chr7_-_81770039
Show fit
|
0.05 |
ENST00000222390.11
ENST00000453411.6
ENST00000457544.7
ENST00000444829.7
|
HGF
|
hepatocyte growth factor
|
|
chr20_+_57561103
Show fit
|
0.05 |
ENST00000319441.6
|
PCK1
|
phosphoenolpyruvate carboxykinase 1
|
|
chr15_-_34583592
Show fit
|
0.05 |
ENST00000683415.1
|
GOLGA8B
|
golgin A8 family member B
|
|
chr17_-_41467386
Show fit
|
0.05 |
ENST00000225899.4
|
KRT32
|
keratin 32
|
|
chr2_-_157444044
Show fit
|
0.04 |
ENST00000264192.8
|
CYTIP
|
cytohesin 1 interacting protein
|
|
chr8_-_7486400
Show fit
|
0.04 |
ENST00000335479.2
|
DEFB106B
|
defensin beta 106B
|
|
chr11_+_118304721
Show fit
|
0.04 |
ENST00000361763.9
|
CD3E
|
CD3e molecule
|
|
chr3_+_122055355
Show fit
|
0.04 |
ENST00000330540.7
ENST00000469710.5
ENST00000493101.5
|
CD86
|
CD86 molecule
|
|
chr7_-_42152444
Show fit
|
0.04 |
ENST00000479210.1
|
GLI3
|
GLI family zinc finger 3
|
|
chr17_-_10469558
Show fit
|
0.04 |
ENST00000255381.2
|
MYH4
|
myosin heavy chain 4
|
|
chr2_+_210477676
Show fit
|
0.04 |
ENST00000673510.1
ENST00000673630.1
ENST00000430249.7
|
CPS1
|
carbamoyl-phosphate synthase 1
|
|
chr2_+_210477708
Show fit
|
0.04 |
ENST00000673711.1
|
CPS1
|
carbamoyl-phosphate synthase 1
|
|
chr8_+_7825137
Show fit
|
0.03 |
ENST00000335186.3
|
DEFB106A
|
defensin beta 106A
|
|
chr12_+_109139397
Show fit
|
0.03 |
ENST00000377854.9
ENST00000377848.7
|
ACACB
|
acetyl-CoA carboxylase beta
|
|
chr3_+_108822759
Show fit
|
0.03 |
ENST00000426646.1
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1
|
|
chr18_+_31318144
Show fit
|
0.03 |
ENST00000257192.5
|
DSG1
|
desmoglein 1
|
|
chr7_-_25228485
Show fit
|
0.03 |
ENST00000222674.2
|
NPVF
|
neuropeptide VF precursor
|
|
chr3_+_63443076
Show fit
|
0.03 |
ENST00000295894.9
|
SYNPR
|
synaptoporin
|
|
chr1_+_40040720
Show fit
|
0.03 |
ENST00000414893.5
ENST00000372792.7
ENST00000372805.8
ENST00000414281.5
ENST00000420216.5
ENST00000372798.5
ENST00000340450.7
ENST00000435719.5
ENST00000427843.5
ENST00000417287.5
ENST00000424977.1
|
CAP1
|
cyclase associated actin cytoskeleton regulatory protein 1
|
|
chr17_-_7929793
Show fit
|
0.03 |
ENST00000303790.3
|
KCNAB3
|
potassium voltage-gated channel subfamily A regulatory beta subunit 3
|
|
chr20_-_51802433
Show fit
|
0.03 |
ENST00000395997.3
|
SALL4
|
spalt like transcription factor 4
|
|
chr3_+_108822778
Show fit
|
0.03 |
ENST00000295756.11
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1
|
|
chr17_+_7407838
Show fit
|
0.03 |
ENST00000302926.7
|
NLGN2
|
neuroligin 2
|
|
chr1_+_50108856
Show fit
|
0.03 |
ENST00000650764.1
ENST00000494555.2
ENST00000371824.7
ENST00000371823.8
ENST00000652693.1
|
ELAVL4
|
ELAV like RNA binding protein 4
|
|
chr1_-_31919093
Show fit
|
0.03 |
ENST00000602683.5
ENST00000470404.5
|
PTP4A2
|
protein tyrosine phosphatase 4A2
|
|
chr16_+_28494634
Show fit
|
0.03 |
ENST00000564831.6
ENST00000431282.2
|
APOBR
|
apolipoprotein B receptor
|
|
chr7_-_42152396
Show fit
|
0.03 |
ENST00000642432.1
ENST00000647255.1
ENST00000677288.1
|
GLI3
|
GLI family zinc finger 3
|
|
chr10_+_93612532
Show fit
|
0.03 |
ENST00000371447.4
|
PDE6C
|
phosphodiesterase 6C
|
|
chr2_-_69643152
Show fit
|
0.02 |
ENST00000606389.7
|
AAK1
|
AP2 associated kinase 1
|
|
chr1_-_193059489
Show fit
|
0.02 |
ENST00000367455.8
ENST00000421683.1
|
UCHL5
|
ubiquitin C-terminal hydrolase L5
|
|
chr17_+_50165990
Show fit
|
0.02 |
ENST00000262018.8
ENST00000344627.10
ENST00000682109.1
ENST00000504073.2
|
SGCA
|
sarcoglycan alpha
|
|
chr19_-_35228699
Show fit
|
0.02 |
ENST00000324675.3
|
FAM187B
|
family with sequence similarity 187 member B
|
|
chr1_+_193059587
Show fit
|
0.02 |
ENST00000400968.7
ENST00000415442.2
ENST00000506303.1
|
RO60
|
Ro60, Y RNA binding protein
|
|
chrX_-_134797134
Show fit
|
0.02 |
ENST00000370790.5
ENST00000493333.5
ENST00000611027.2
ENST00000343004.10
ENST00000298090.10
|
PABIR2
|
PABIR family member 2
|
|
chr1_-_182671853
Show fit
|
0.02 |
ENST00000367556.5
|
RGS8
|
regulator of G protein signaling 8
|
|
chrX_+_69616067
Show fit
|
0.02 |
ENST00000338901.4
ENST00000525810.5
ENST00000527388.5
ENST00000374553.6
ENST00000374552.9
ENST00000524573.5
|
EDA
|
ectodysplasin A
|
|
chr1_+_193059448
Show fit
|
0.02 |
ENST00000432079.5
|
RO60
|
Ro60, Y RNA binding protein
|
|
chr1_-_182671902
Show fit
|
0.02 |
ENST00000483095.6
|
RGS8
|
regulator of G protein signaling 8
|
|
chr17_-_10026265
Show fit
|
0.02 |
ENST00000437099.6
ENST00000396115.6
|
GAS7
|
growth arrest specific 7
|
|
chr17_-_81656532
Show fit
|
0.02 |
ENST00000331056.10
|
PDE6G
|
phosphodiesterase 6G
|
|
chr6_+_118548289
Show fit
|
0.02 |
ENST00000357525.6
|
PLN
|
phospholamban
|
|
chrX_+_103585478
Show fit
|
0.01 |
ENST00000468024.5
ENST00000415568.2
ENST00000472484.6
ENST00000613326.4
ENST00000490644.5
ENST00000459722.1
ENST00000472745.1
ENST00000494801.5
ENST00000434216.2
|
TCEAL4
|
transcription elongation factor A like 4
|
|
chr14_-_70416994
Show fit
|
0.01 |
ENST00000621525.4
ENST00000256366.6
|
SYNJ2BP-COX16
SYNJ2BP
|
SYNJ2BP-COX16 readthrough
synaptojanin 2 binding protein
|
|
chr6_-_138987640
Show fit
|
0.01 |
ENST00000529597.5
ENST00000415951.6
ENST00000367663.8
ENST00000409812.6
|
REPS1
|
RALBP1 associated Eps domain containing 1
|
|
chr6_-_73521783
Show fit
|
0.01 |
ENST00000331523.7
ENST00000356303.7
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr11_-_31509588
Show fit
|
0.01 |
ENST00000534812.5
ENST00000529749.5
ENST00000532287.6
ENST00000278200.5
ENST00000530023.5
ENST00000533642.1
|
IMMP1L
|
inner mitochondrial membrane peptidase subunit 1
|
|
chr10_+_97446137
Show fit
|
0.01 |
ENST00000370854.7
ENST00000393760.6
ENST00000414567.5
|
ZDHHC16
|
zinc finger DHHC-type palmitoyltransferase 16
|
|
chr6_-_111483700
Show fit
|
0.01 |
ENST00000435970.5
ENST00000358835.7
|
REV3L
|
REV3 like, DNA directed polymerase zeta catalytic subunit
|
|
chr10_+_18260715
Show fit
|
0.01 |
ENST00000615785.4
ENST00000617363.4
ENST00000396576.6
|
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2
|
|
chr17_+_58169401
Show fit
|
0.01 |
ENST00000641866.1
|
OR4D2
|
olfactory receptor family 4 subfamily D member 2
|
|
chr12_+_53454819
Show fit
|
0.01 |
ENST00000562264.5
|
PCBP2
|
poly(rC) binding protein 2
|
|
chr10_+_97446194
Show fit
|
0.01 |
ENST00000370846.8
ENST00000352634.8
ENST00000353979.7
ENST00000370842.6
ENST00000345745.9
|
ZDHHC16
|
zinc finger DHHC-type palmitoyltransferase 16
|
|
chr10_-_97445850
Show fit
|
0.00 |
ENST00000477692.6
ENST00000485122.6
ENST00000370886.9
ENST00000370885.8
ENST00000370902.8
ENST00000370884.5
|
EXOSC1
|
exosome component 1
|
|
chr12_+_53454764
Show fit
|
0.00 |
ENST00000439930.7
ENST00000548933.5
|
PCBP2
|
poly(rC) binding protein 2
|
|
chr12_+_56116623
Show fit
|
0.00 |
ENST00000546591.6
|
RPL41
|
ribosomal protein L41
|
|
chr12_+_53454718
Show fit
|
0.00 |
ENST00000552819.5
ENST00000455667.7
|
PCBP2
|
poly(rC) binding protein 2
|
|
chr12_+_53954870
Show fit
|
0.00 |
ENST00000243103.4
|
HOXC12
|
homeobox C12
|
|
chr12_+_56116584
Show fit
|
0.00 |
ENST00000501597.3
|
RPL41
|
ribosomal protein L41
|
|
chr1_-_244862381
Show fit
|
0.00 |
ENST00000640001.1
ENST00000639628.1
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U
|