Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
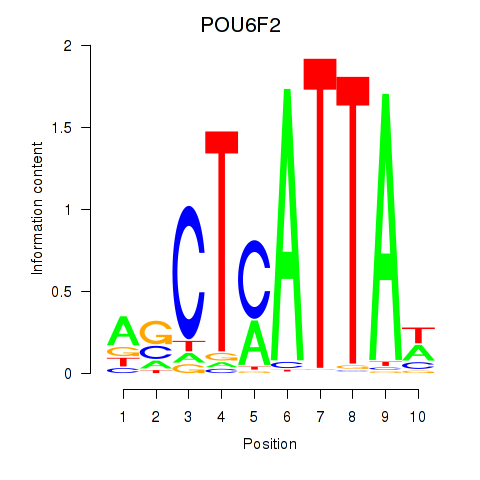
Results for POU6F2
Z-value: 0.46
Transcription factors associated with POU6F2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU6F2
|
ENSG00000106536.21 | POU6F2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU6F2 | hg38_v1_chr7_+_38977904_38978009 | -0.11 | 5.9e-01 | Click! |
Activity profile of POU6F2 motif
Sorted Z-values of POU6F2 motif
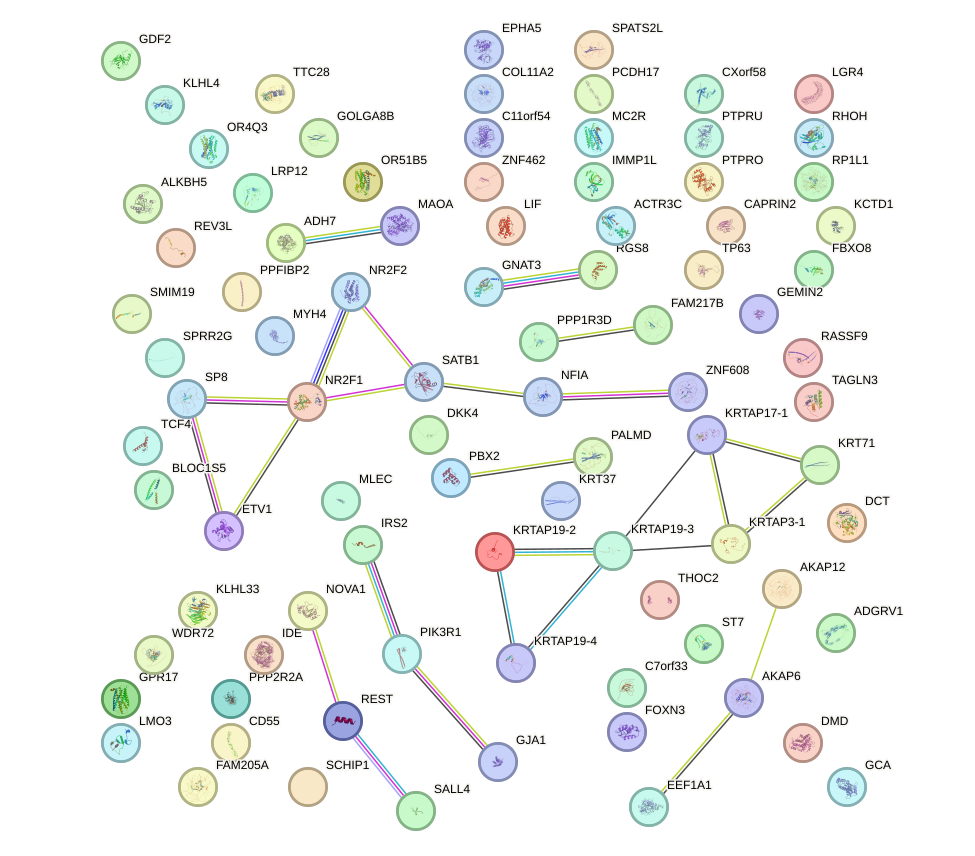
Network of associatons between targets according to the STRING database.
First level regulatory network of POU6F2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_57631735 | 1.08 |
ENST00000377918.8
|
PCDH17
|
protocadherin 17 |
| chr12_-_30735014 | 1.02 |
ENST00000433722.6
|
CAPRIN2
|
caprin family member 2 |
| chr20_+_59940362 | 0.87 |
ENST00000360816.8
|
FAM217B
|
family with sequence similarity 217 member B |
| chr16_+_53099100 | 0.64 |
ENST00000565832.5
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr1_-_182672232 | 0.63 |
ENST00000508450.5
|
RGS8
|
regulator of G protein signaling 8 |
| chr11_+_7576975 | 0.59 |
ENST00000684215.1
ENST00000650027.1 |
PPFIBP2
|
PPFIA binding protein 2 |
| chr2_+_200440649 | 0.56 |
ENST00000366118.2
|
SPATS2L
|
spermatogenesis associated serine rich 2 like |
| chr1_+_99646025 | 0.49 |
ENST00000263174.9
ENST00000605497.5 ENST00000615664.1 |
PALMD
|
palmdelphin |
| chr21_-_30487436 | 0.45 |
ENST00000334055.5
|
KRTAP19-2
|
keratin associated protein 19-2 |
| chr12_-_85836372 | 0.45 |
ENST00000361228.5
|
RASSF9
|
Ras association domain family member 9 |
| chr6_-_111483700 | 0.43 |
ENST00000435970.5
ENST00000358835.7 |
REV3L
|
REV3 like, DNA directed polymerase zeta catalytic subunit |
| chr6_-_8064333 | 0.42 |
ENST00000543936.7
ENST00000397457.7 |
BLOC1S5
|
biogenesis of lysosomal organelles complex 1 subunit 5 |
| chr15_+_96325935 | 0.42 |
ENST00000421109.6
|
NR2F2
|
nuclear receptor subfamily 2 group F member 2 |
| chr20_-_59940289 | 0.40 |
ENST00000370996.5
|
PPP1R3D
|
protein phosphatase 1 regulatory subunit 3D |
| chr2_-_70553440 | 0.39 |
ENST00000450929.5
|
TGFA
|
transforming growth factor alpha |
| chrX_-_32412220 | 0.36 |
ENST00000619831.5
|
DMD
|
dystrophin |
| chr18_-_26549402 | 0.35 |
ENST00000408011.7
|
KCTD1
|
potassium channel tetramerization domain containing 1 |
| chr7_-_20786879 | 0.33 |
ENST00000418710.3
ENST00000617581.4 ENST00000361443.4 |
SP8
|
Sp8 transcription factor |
| chr14_-_26598025 | 0.32 |
ENST00000539517.7
|
NOVA1
|
NOVA alternative splicing regulator 1 |
| chr17_+_18183052 | 0.32 |
ENST00000541285.1
|
ALKBH5
|
alkB homolog 5, RNA demethylase |
| chr6_+_130018565 | 0.32 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chr21_-_30497160 | 0.31 |
ENST00000334058.3
|
KRTAP19-4
|
keratin associated protein 19-4 |
| chr14_+_32329256 | 0.31 |
ENST00000280979.9
|
AKAP6
|
A-kinase anchoring protein 6 |
| chr6_+_121437378 | 0.30 |
ENST00000650427.1
ENST00000647564.1 |
GJA1
|
gap junction protein alpha 1 |
| chr3_+_159839847 | 0.29 |
ENST00000445224.6
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr9_+_114611206 | 0.29 |
ENST00000374049.4
ENST00000288502.9 |
TMEM268
|
transmembrane protein 268 |
| chr3_-_18425295 | 0.29 |
ENST00000338745.11
ENST00000450898.1 |
SATB1
|
SATB homeobox 1 |
| chr4_+_40196907 | 0.27 |
ENST00000622175.4
ENST00000619474.4 ENST00000615083.4 ENST00000610353.4 ENST00000614836.1 |
RHOH
|
ras homolog family member H |
| chr4_+_56907876 | 0.26 |
ENST00000640168.2
ENST00000309042.12 |
REST
|
RE1 silencing transcription factor |
| chr17_+_18183803 | 0.26 |
ENST00000399138.5
|
ALKBH5
|
alkB homolog 5, RNA demethylase |
| chr17_-_41315706 | 0.26 |
ENST00000334202.5
|
KRTAP17-1
|
keratin associated protein 17-1 |
| chr14_+_32329341 | 0.26 |
ENST00000557354.5
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A-kinase anchoring protein 6 |
| chr6_+_157036315 | 0.25 |
ENST00000637904.1
|
ARID1B
|
AT-rich interaction domain 1B |
| chr13_-_109786567 | 0.25 |
ENST00000375856.5
|
IRS2
|
insulin receptor substrate 2 |
| chr12_+_15546344 | 0.24 |
ENST00000674388.1
ENST00000542557.5 ENST00000445537.6 ENST00000544244.5 ENST00000442921.7 |
PTPRO
|
protein tyrosine phosphatase receptor type O |
| chr4_+_40197023 | 0.24 |
ENST00000381799.10
|
RHOH
|
ras homolog family member H |
| chr17_-_41424583 | 0.24 |
ENST00000225550.4
|
KRT37
|
keratin 37 |
| chr9_+_106863121 | 0.24 |
ENST00000472574.1
ENST00000277225.10 |
ZNF462
|
zinc finger protein 462 |
| chr11_-_31509569 | 0.23 |
ENST00000526776.5
|
IMMP1L
|
inner mitochondrial membrane peptidase subunit 1 |
| chr11_-_27472698 | 0.22 |
ENST00000389858.4
ENST00000379214.9 |
LGR4
|
leucine rich repeat containing G protein-coupled receptor 4 |
| chr15_-_53733103 | 0.22 |
ENST00000559418.5
|
WDR72
|
WD repeat domain 72 |
| chr3_+_111999326 | 0.20 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr21_-_30492008 | 0.20 |
ENST00000334063.6
|
KRTAP19-3
|
keratin associated protein 19-3 |
| chr1_-_153150884 | 0.19 |
ENST00000368748.5
|
SPRR2G
|
small proline rich protein 2G |
| chr5_+_68290637 | 0.19 |
ENST00000336483.9
|
PIK3R1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr1_+_207325629 | 0.18 |
ENST00000618707.2
|
CD55
|
CD55 molecule (Cromer blood group) |
| chr2_+_162318884 | 0.18 |
ENST00000446271.5
ENST00000429691.6 |
GCA
|
grancalcin |
| chr3_+_189789672 | 0.17 |
ENST00000434928.5
|
TP63
|
tumor protein p63 |
| chr11_-_31509588 | 0.17 |
ENST00000534812.5
ENST00000529749.5 ENST00000532287.6 ENST00000278200.5 ENST00000530023.5 ENST00000533642.1 |
IMMP1L
|
inner mitochondrial membrane peptidase subunit 1 |
| chr14_-_20436161 | 0.17 |
ENST00000636854.3
|
KLHL33
|
kelch like family member 33 |
| chr5_+_90640718 | 0.16 |
ENST00000640403.1
|
ADGRV1
|
adhesion G protein-coupled receptor V1 |
| chr18_-_13915531 | 0.16 |
ENST00000327606.4
|
MC2R
|
melanocortin 2 receptor |
| chr6_+_29306626 | 0.16 |
ENST00000377160.4
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1 |
| chr1_+_61404076 | 0.15 |
ENST00000357977.5
|
NFIA
|
nuclear factor I A |
| chr1_-_193059296 | 0.15 |
ENST00000367450.7
ENST00000367451.8 ENST00000367454.6 ENST00000367448.5 ENST00000367449.5 |
UCHL5
|
ubiquitin C-terminal hydrolase L5 |
| chr5_-_124746630 | 0.14 |
ENST00000513986.2
|
ZNF608
|
zinc finger protein 608 |
| chr7_-_150302980 | 0.14 |
ENST00000252071.8
|
ACTR3C
|
actin related protein 3C |
| chr7_-_80512041 | 0.14 |
ENST00000398291.4
|
GNAT3
|
G protein subunit alpha transducin 3 |
| chr20_-_51802433 | 0.13 |
ENST00000395997.3
|
SALL4
|
spalt like transcription factor 4 |
| chr8_+_26293112 | 0.13 |
ENST00000523925.5
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2 regulatory subunit Balpha |
| chr8_-_42501224 | 0.13 |
ENST00000520262.6
ENST00000517366.1 |
SLC20A2
|
solute carrier family 20 member 2 |
| chr1_-_193059489 | 0.13 |
ENST00000367455.8
ENST00000421683.1 |
UCHL5
|
ubiquitin C-terminal hydrolase L5 |
| chr17_-_41009124 | 0.12 |
ENST00000391588.3
|
KRTAP3-1
|
keratin associated protein 3-1 |
| chr14_+_39114289 | 0.12 |
ENST00000396249.7
ENST00000250379.13 ENST00000534684.7 ENST00000308317.12 ENST00000527381.2 |
GEMIN2
|
gem nuclear organelle associated protein 2 |
| chr14_-_89619118 | 0.12 |
ENST00000345097.8
ENST00000555855.5 ENST00000555353.5 |
FOXN3
|
forkhead box N3 |
| chr6_-_33192454 | 0.12 |
ENST00000395194.1
ENST00000341947.7 ENST00000374708.8 |
COL11A2
|
collagen type XI alpha 2 chain |
| chr5_+_135579193 | 0.11 |
ENST00000646290.1
|
SLC25A48
|
solute carrier family 25 member 48 |
| chr7_+_148590760 | 0.11 |
ENST00000307003.3
|
C7orf33
|
chromosome 7 open reading frame 33 |
| chr6_+_151325665 | 0.11 |
ENST00000354675.10
|
AKAP12
|
A-kinase anchoring protein 12 |
| chr7_+_117020191 | 0.11 |
ENST00000434836.5
ENST00000393443.5 ENST00000465133.5 ENST00000477742.5 ENST00000393444.7 ENST00000393447.8 |
ST7
|
suppression of tumorigenicity 7 |
| chr10_+_47322450 | 0.11 |
ENST00000581492.3
|
GDF2
|
growth differentiation factor 2 |
| chr8_-_42377227 | 0.11 |
ENST00000220812.3
|
DKK4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr9_-_34729482 | 0.11 |
ENST00000378788.4
|
FAM205A
|
family with sequence similarity 205 member A |
| chr22_-_30246739 | 0.11 |
ENST00000403987.3
ENST00000249075.4 |
LIF
|
LIF interleukin 6 family cytokine |
| chr14_+_19743571 | 0.11 |
ENST00000642117.2
|
OR4Q3
|
olfactory receptor family 4 subfamily Q member 3 |
| chr4_-_174283614 | 0.10 |
ENST00000513696.1
ENST00000393674.7 ENST00000503293.5 |
FBXO8
|
F-box protein 8 |
| chr3_-_33645433 | 0.10 |
ENST00000635664.1
ENST00000485378.6 ENST00000313350.10 ENST00000487200.5 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr8_+_42541128 | 0.10 |
ENST00000438528.7
|
SMIM19
|
small integral membrane protein 19 |
| chrX_-_123622809 | 0.10 |
ENST00000441692.5
|
THOC2
|
THO complex 2 |
| chr22_-_28306645 | 0.10 |
ENST00000612946.4
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr12_-_16606102 | 0.09 |
ENST00000537304.6
|
LMO3
|
LIM domain only 3 |
| chrX_-_123623155 | 0.09 |
ENST00000618150.4
|
THOC2
|
THO complex 2 |
| chr12_-_16605939 | 0.09 |
ENST00000541295.5
ENST00000535535.5 |
LMO3
|
LIM domain only 3 |
| chr10_-_92497727 | 0.09 |
ENST00000496903.5
ENST00000678824.1 ENST00000371581.9 |
IDE
|
insulin degrading enzyme |
| chr7_-_13988863 | 0.08 |
ENST00000405358.8
|
ETV1
|
ETS variant transcription factor 1 |
| chrX_+_87517784 | 0.08 |
ENST00000373119.9
ENST00000373114.4 |
KLHL4
|
kelch like family member 4 |
| chrX_+_43656289 | 0.08 |
ENST00000338702.4
|
MAOA
|
monoamine oxidase A |
| chr11_+_93746433 | 0.08 |
ENST00000526335.1
|
C11orf54
|
chromosome 11 open reading frame 54 |
| chr6_-_73521783 | 0.08 |
ENST00000331523.7
ENST00000356303.7 |
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr6_-_32190170 | 0.08 |
ENST00000375050.6
|
PBX2
|
PBX homeobox 2 |
| chr8_-_10655137 | 0.07 |
ENST00000382483.4
|
RP1L1
|
RP1 like 1 |
| chrX_+_23908006 | 0.07 |
ENST00000379211.8
ENST00000648352.1 |
CXorf58
|
chromosome X open reading frame 58 |
| chr12_-_52553139 | 0.07 |
ENST00000267119.6
|
KRT71
|
keratin 71 |
| chr15_-_34583592 | 0.07 |
ENST00000683415.1
|
GOLGA8B
|
golgin A8 family member B |
| chr11_-_5343524 | 0.07 |
ENST00000300773.3
|
OR51B5
|
olfactory receptor family 51 subfamily B member 5 |
| chr18_-_55423757 | 0.06 |
ENST00000675707.1
|
TCF4
|
transcription factor 4 |
| chr4_-_99144238 | 0.06 |
ENST00000512499.5
ENST00000504125.1 ENST00000505590.5 ENST00000629236.2 ENST00000508393.5 ENST00000265512.12 |
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr4_-_65670478 | 0.06 |
ENST00000613740.5
ENST00000622150.4 ENST00000511294.1 |
EPHA5
|
EPH receptor A5 |
| chr12_+_120694167 | 0.06 |
ENST00000535656.1
|
MLEC
|
malectin |
| chr17_-_10469558 | 0.06 |
ENST00000255381.2
|
MYH4
|
myosin heavy chain 4 |
| chr4_-_65670339 | 0.06 |
ENST00000273854.7
|
EPHA5
|
EPH receptor A5 |
| chr14_+_22515623 | 0.06 |
ENST00000390509.1
|
TRAJ28
|
T cell receptor alpha joining 28 |
| chr13_-_94479671 | 0.05 |
ENST00000377028.10
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr2_+_127646145 | 0.05 |
ENST00000486700.2
ENST00000272644.7 |
GPR17
|
G protein-coupled receptor 17 |
| chr13_-_44161257 | 0.05 |
ENST00000400419.2
|
SMIM2
|
small integral membrane protein 2 |
| chr7_+_129368123 | 0.05 |
ENST00000460109.5
ENST00000474594.5 |
AHCYL2
|
adenosylhomocysteinase like 2 |
| chr21_-_30881572 | 0.05 |
ENST00000332378.6
|
KRTAP11-1
|
keratin associated protein 11-1 |
| chr5_+_36606355 | 0.05 |
ENST00000681909.1
ENST00000513903.5 ENST00000681795.1 ENST00000680125.1 ENST00000612708.5 ENST00000680232.1 ENST00000681776.1 ENST00000681926.1 ENST00000679958.1 ENST00000265113.9 ENST00000504121.5 ENST00000512374.1 ENST00000613445.5 ENST00000679983.1 |
SLC1A3
|
solute carrier family 1 member 3 |
| chr17_-_42185452 | 0.05 |
ENST00000293330.1
|
HCRT
|
hypocretin neuropeptide precursor |
| chr6_-_110815152 | 0.05 |
ENST00000413605.6
|
CDK19
|
cyclin dependent kinase 19 |
| chr12_-_16608073 | 0.04 |
ENST00000441439.6
|
LMO3
|
LIM domain only 3 |
| chr10_-_126521439 | 0.04 |
ENST00000284694.11
ENST00000432642.5 |
C10orf90
|
chromosome 10 open reading frame 90 |
| chr7_-_126533850 | 0.04 |
ENST00000444921.3
|
GRM8
|
glutamate metabotropic receptor 8 |
| chr2_-_2324323 | 0.04 |
ENST00000648339.1
ENST00000647694.1 |
MYT1L
|
myelin transcription factor 1 like |
| chr1_+_175067831 | 0.04 |
ENST00000239462.9
|
TNN
|
tenascin N |
| chr19_-_14475307 | 0.04 |
ENST00000292513.4
|
PTGER1
|
prostaglandin E receptor 1 |
| chr4_+_112860912 | 0.04 |
ENST00000671951.1
|
ANK2
|
ankyrin 2 |
| chr1_-_10964201 | 0.04 |
ENST00000418570.6
|
C1orf127
|
chromosome 1 open reading frame 127 |
| chr7_+_129375643 | 0.04 |
ENST00000490911.5
|
AHCYL2
|
adenosylhomocysteinase like 2 |
| chr7_-_13989658 | 0.03 |
ENST00000430479.6
ENST00000433547.1 ENST00000405192.6 |
ETV1
|
ETS variant transcription factor 1 |
| chr9_-_76906041 | 0.03 |
ENST00000443509.6
ENST00000428286.5 ENST00000376713.3 |
PRUNE2
|
prune homolog 2 with BCH domain |
| chr4_+_112861053 | 0.03 |
ENST00000672221.1
|
ANK2
|
ankyrin 2 |
| chr4_+_112860981 | 0.03 |
ENST00000671704.1
|
ANK2
|
ankyrin 2 |
| chr4_+_85604146 | 0.03 |
ENST00000512201.5
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr6_-_110815408 | 0.03 |
ENST00000368911.8
|
CDK19
|
cyclin dependent kinase 19 |
| chr3_+_189789643 | 0.03 |
ENST00000354600.10
|
TP63
|
tumor protein p63 |
| chr12_-_16608183 | 0.03 |
ENST00000354662.5
ENST00000538051.5 |
LMO3
|
LIM domain only 3 |
| chr2_+_127645864 | 0.03 |
ENST00000544369.5
|
GPR17
|
G protein-coupled receptor 17 |
| chr2_-_68871382 | 0.03 |
ENST00000295379.2
|
BMP10
|
bone morphogenetic protein 10 |
| chr16_-_18926408 | 0.03 |
ENST00000446231.7
|
SMG1
|
SMG1 nonsense mediated mRNA decay associated PI3K related kinase |
| chr15_+_58410543 | 0.03 |
ENST00000356113.10
ENST00000414170.7 |
LIPC
|
lipase C, hepatic type |
| chr2_+_44275473 | 0.03 |
ENST00000260649.11
|
SLC3A1
|
solute carrier family 3 member 1 |
| chr11_+_107591077 | 0.02 |
ENST00000531234.5
ENST00000265840.12 |
ELMOD1
|
ELMO domain containing 1 |
| chr9_-_76906090 | 0.02 |
ENST00000376718.8
|
PRUNE2
|
prune homolog 2 with BCH domain |
| chrX_+_108045050 | 0.02 |
ENST00000458383.1
ENST00000217957.10 |
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr1_-_182671853 | 0.02 |
ENST00000367556.5
|
RGS8
|
regulator of G protein signaling 8 |
| chr10_-_121596117 | 0.02 |
ENST00000351936.11
|
FGFR2
|
fibroblast growth factor receptor 2 |
| chr14_-_36519679 | 0.02 |
ENST00000498187.6
|
NKX2-1
|
NK2 homeobox 1 |
| chrX_+_108044967 | 0.02 |
ENST00000415430.7
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr6_+_25754699 | 0.02 |
ENST00000439485.6
ENST00000377905.9 |
SLC17A4
|
solute carrier family 17 member 4 |
| chr19_+_9178979 | 0.02 |
ENST00000642043.1
ENST00000641288.2 |
OR7D2
|
olfactory receptor family 7 subfamily D member 2 |
| chr1_-_182671902 | 0.02 |
ENST00000483095.6
|
RGS8
|
regulator of G protein signaling 8 |
| chr2_-_2326378 | 0.02 |
ENST00000647618.1
|
MYT1L
|
myelin transcription factor 1 like |
| chr1_+_45913647 | 0.02 |
ENST00000674079.1
|
MAST2
|
microtubule associated serine/threonine kinase 2 |
| chr7_-_73624492 | 0.01 |
ENST00000414749.6
ENST00000429400.6 ENST00000434326.5 ENST00000313375.8 ENST00000354613.5 ENST00000453275.1 |
MLXIPL
|
MLX interacting protein like |
| chr12_-_52652207 | 0.01 |
ENST00000309680.4
|
KRT2
|
keratin 2 |
| chr2_-_55917699 | 0.01 |
ENST00000634374.1
|
EFEMP1
|
EGF containing fibulin extracellular matrix protein 1 |
| chr3_+_111999189 | 0.01 |
ENST00000455401.6
|
TAGLN3
|
transgelin 3 |
| chr17_+_7407838 | 0.01 |
ENST00000302926.7
|
NLGN2
|
neuroligin 2 |
| chr17_+_39628496 | 0.01 |
ENST00000394265.5
ENST00000394267.2 |
PPP1R1B
|
protein phosphatase 1 regulatory inhibitor subunit 1B |
| chr13_+_45464901 | 0.01 |
ENST00000349995.10
|
COG3
|
component of oligomeric golgi complex 3 |
| chr3_+_111998915 | 0.01 |
ENST00000478951.6
|
TAGLN3
|
transgelin 3 |
| chr11_+_71833200 | 0.01 |
ENST00000328698.2
|
DEFB108B
|
defensin beta 108B |
| chr11_-_26572102 | 0.01 |
ENST00000455601.6
|
MUC15
|
mucin 15, cell surface associated |
| chr3_-_101320558 | 0.01 |
ENST00000193391.8
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2 |
| chr6_+_50818857 | 0.01 |
ENST00000393655.4
|
TFAP2B
|
transcription factor AP-2 beta |
| chr2_+_168901290 | 0.01 |
ENST00000429379.2
ENST00000375363.8 ENST00000421979.1 |
G6PC2
|
glucose-6-phosphatase catalytic subunit 2 |
| chrX_-_155071064 | 0.00 |
ENST00000369484.8
ENST00000369476.8 |
CMC4
MTCP1
|
C-X9-C motif containing 4 mature T cell proliferation 1 |
| chr7_+_116222804 | 0.00 |
ENST00000393481.6
|
TES
|
testin LIM domain protein |
| chr2_-_157444044 | 0.00 |
ENST00000264192.8
|
CYTIP
|
cytohesin 1 interacting protein |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 | 0.4 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 0.3 | GO:0097086 | amniotic stem cell differentiation(GO:0097086) negative regulation of dense core granule biogenesis(GO:2000706) negative regulation of mesenchymal stem cell differentiation(GO:2000740) regulation of amniotic stem cell differentiation(GO:2000797) negative regulation of amniotic stem cell differentiation(GO:2000798) |
| 0.1 | 0.6 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.2 | GO:0061290 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.1 | 0.3 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.1 | 0.2 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 1.1 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.0 | 0.3 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.4 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.3 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.7 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.3 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.0 | 0.2 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.0 | 0.4 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.4 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.4 | GO:0050942 | positive regulation of pigment cell differentiation(GO:0050942) |
| 0.0 | 0.3 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 1.0 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.3 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.3 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.4 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.6 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 0.4 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.4 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.4 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.4 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.7 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.3 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |