Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
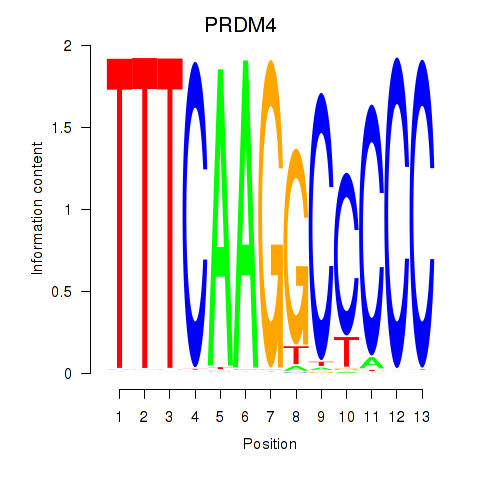
Results for PRDM4
Z-value: 0.34
Transcription factors associated with PRDM4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRDM4
|
ENSG00000110851.12 | PRDM4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PRDM4 | hg38_v1_chr12_-_107761113_107761176 | 0.29 | 1.6e-01 | Click! |
Activity profile of PRDM4 motif
Sorted Z-values of PRDM4 motif
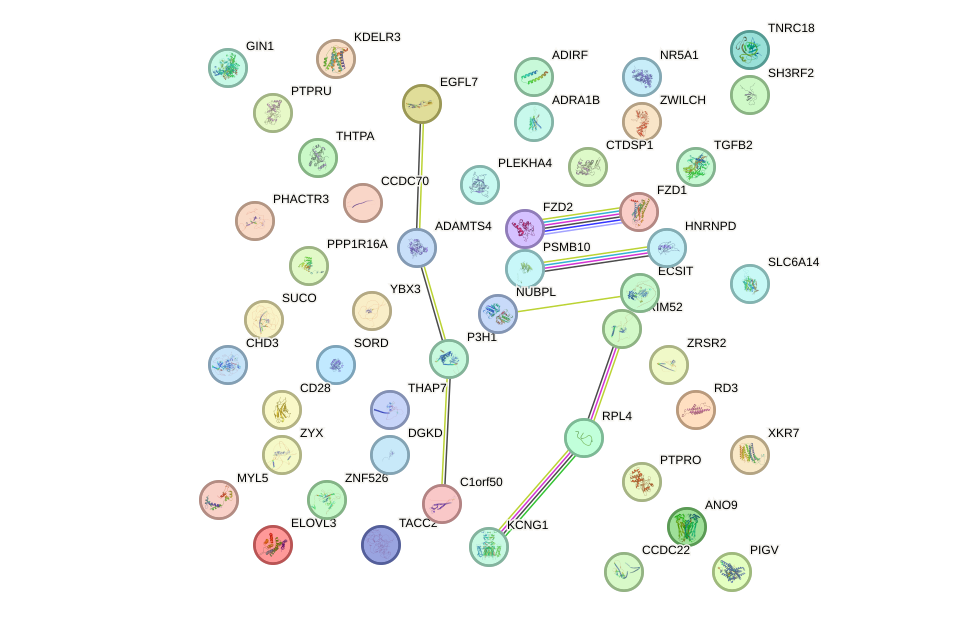
Network of associatons between targets according to the STRING database.
First level regulatory network of PRDM4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_124500986 | 0.73 |
ENST00000373587.3
|
NR5A1
|
nuclear receptor subfamily 5 group A member 1 |
| chr11_-_441964 | 0.66 |
ENST00000332826.7
|
ANO9
|
anoctamin 9 |
| chr15_-_35085295 | 0.53 |
ENST00000528386.4
|
NANOGP8
|
Nanog homeobox retrogene P8 |
| chr16_-_67936808 | 0.52 |
ENST00000358514.9
|
PSMB10
|
proteasome 20S subunit beta 10 |
| chr19_+_42220283 | 0.51 |
ENST00000301215.8
ENST00000597945.1 |
ZNF526
|
zinc finger protein 526 |
| chr7_+_143381907 | 0.36 |
ENST00000392910.6
|
ZYX
|
zyxin |
| chr7_+_143381561 | 0.35 |
ENST00000354434.8
|
ZYX
|
zyxin |
| chr8_+_144477975 | 0.31 |
ENST00000435887.2
|
PPP1R16A
|
protein phosphatase 1 regulatory subunit 16A |
| chr10_+_86968432 | 0.22 |
ENST00000416348.1
ENST00000372013.8 |
ADIRF
|
adipogenesis regulatory factor |
| chr1_+_172533104 | 0.22 |
ENST00000616058.4
ENST00000263688.4 ENST00000610051.5 |
SUCO
|
SUN domain containing ossification factor |
| chr19_-_48868454 | 0.21 |
ENST00000355496.9
|
PLEKHA4
|
pleckstrin homology domain containing A4 |
| chr7_-_5423826 | 0.20 |
ENST00000430969.6
|
TNRC18
|
trinucleotide repeat containing 18 |
| chr10_+_102226293 | 0.19 |
ENST00000370005.4
|
ELOVL3
|
ELOVL fatty acid elongase 3 |
| chr19_-_48868590 | 0.19 |
ENST00000263265.11
|
PLEKHA4
|
pleckstrin homology domain containing A4 |
| chr17_-_35487739 | 0.18 |
ENST00000628453.3
|
ENSG00000205045.11
|
schlafen family member 12 like |
| chr1_+_171781635 | 0.17 |
ENST00000361735.4
ENST00000362019.7 ENST00000367737.9 |
EEF1AKNMT
|
eEF1A lysine and N-terminal methyltransferase |
| chr2_+_218399838 | 0.17 |
ENST00000273062.7
|
CTDSP1
|
CTD small phosphatase 1 |
| chr6_-_32192630 | 0.16 |
ENST00000375040.8
|
GPSM3
|
G protein signaling modulator 3 |
| chr20_-_51023081 | 0.16 |
ENST00000433903.5
ENST00000424171.5 ENST00000371571.5 ENST00000439216.5 |
KCNG1
|
potassium voltage-gated channel modifier subfamily G member 1 |
| chr20_+_31968141 | 0.16 |
ENST00000562532.3
|
XKR7
|
XK related 7 |
| chr7_+_91264426 | 0.15 |
ENST00000287934.4
|
FZD1
|
frizzled class receptor 1 |
| chr5_-_103120097 | 0.14 |
ENST00000508629.5
ENST00000399004.7 |
GIN1
|
gypsy retrotransposon integrase 1 |
| chr10_+_122163590 | 0.14 |
ENST00000368999.5
|
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chrX_+_15790446 | 0.13 |
ENST00000380308.7
ENST00000307771.8 |
ZRSR2
|
zinc finger CCCH-type, RNA binding motif and serine/arginine rich 2 |
| chr6_-_32192845 | 0.13 |
ENST00000487761.5
|
GPSM3
|
G protein signaling modulator 3 |
| chr1_+_6451304 | 0.13 |
ENST00000636644.1
|
ESPN
|
espin |
| chr13_+_51861963 | 0.12 |
ENST00000242819.7
|
CCDC70
|
coiled-coil domain containing 70 |
| chr12_-_10723307 | 0.12 |
ENST00000279550.11
ENST00000228251.9 |
YBX3
|
Y-box binding protein 3 |
| chr5_-_181261078 | 0.10 |
ENST00000611618.1
|
TRIM52
|
tripartite motif containing 52 |
| chr1_-_211492111 | 0.10 |
ENST00000367002.5
ENST00000680073.1 |
RD3
|
RD3 regulator of GUCY2D |
| chr14_+_23555983 | 0.08 |
ENST00000404535.3
|
THTPA
|
thiamine triphosphatase |
| chr22_-_21002081 | 0.08 |
ENST00000215742.9
ENST00000399133.2 |
THAP7
|
THAP domain containing 7 |
| chrX_+_49235460 | 0.07 |
ENST00000376227.4
|
CCDC22
|
coiled-coil domain containing 22 |
| chr1_-_161199044 | 0.07 |
ENST00000367995.3
ENST00000367996.6 |
ADAMTS4
|
ADAM metallopeptidase with thrombospondin type 1 motif 4 |
| chr12_+_15322480 | 0.07 |
ENST00000674188.1
ENST00000281171.9 ENST00000543886.6 |
PTPRO
|
protein tyrosine phosphatase receptor type O |
| chr22_+_38468036 | 0.06 |
ENST00000409006.3
ENST00000216014.9 |
KDELR3
|
KDEL endoplasmic reticulum protein retention receptor 3 |
| chr1_-_42766978 | 0.06 |
ENST00000372526.2
ENST00000236040.8 ENST00000296388.10 ENST00000397054.7 |
P3H1
|
prolyl 3-hydroxylase 1 |
| chr14_+_31561376 | 0.06 |
ENST00000550649.5
ENST00000281081.12 |
NUBPL
|
nucleotide binding protein like |
| chr1_+_6451578 | 0.05 |
ENST00000434576.2
ENST00000477679.2 |
ESPN
|
espin |
| chr2_+_233388146 | 0.05 |
ENST00000409813.7
|
DGKD
|
diacylglycerol kinase delta |
| chr9_+_136665745 | 0.04 |
ENST00000371698.3
|
EGFL7
|
EGF like domain multiple 7 |
| chr1_+_26788166 | 0.03 |
ENST00000374145.6
ENST00000431541.6 ENST00000674273.1 |
PIGV
|
phosphatidylinositol glycan anchor biosynthesis class V |
| chr1_+_42767241 | 0.03 |
ENST00000372525.7
|
C1orf50
|
chromosome 1 open reading frame 50 |
| chr4_-_82373946 | 0.03 |
ENST00000352301.8
ENST00000509107.1 ENST00000353341.8 ENST00000313899.12 |
HNRNPD
|
heterogeneous nuclear ribonucleoprotein D |
| chr19_-_11529094 | 0.03 |
ENST00000588998.5
ENST00000586149.1 |
ECSIT
|
ECSIT signaling integrator |
| chr21_+_33403391 | 0.03 |
ENST00000290219.11
ENST00000381995.5 |
IFNGR2
|
interferon gamma receptor 2 |
| chr15_-_66504832 | 0.02 |
ENST00000569438.2
ENST00000569696.5 ENST00000307961.11 |
RPL4
|
ribosomal protein L4 |
| chrX_+_116436599 | 0.02 |
ENST00000598581.3
|
SLC6A14
|
solute carrier family 6 member 14 |
| chr1_+_26787667 | 0.02 |
ENST00000674335.1
|
PIGV
|
phosphatidylinositol glycan anchor biosynthesis class V |
| chr17_+_44557476 | 0.02 |
ENST00000315323.5
|
FZD2
|
frizzled class receptor 2 |
| chr4_+_673518 | 0.02 |
ENST00000506838.5
|
MYL5
|
myosin light chain 5 |
| chr1_+_26787926 | 0.01 |
ENST00000674202.1
ENST00000674222.1 |
PIGV
|
phosphatidylinositol glycan anchor biosynthesis class V |
| chr5_+_159916475 | 0.01 |
ENST00000306675.5
|
ADRA1B
|
adrenoceptor alpha 1B |
| chr15_+_66504959 | 0.01 |
ENST00000535141.6
ENST00000613446.4 ENST00000446801.6 |
ZWILCH
|
zwilch kinetochore protein |
| chr12_+_15322529 | 0.01 |
ENST00000348962.7
|
PTPRO
|
protein tyrosine phosphatase receptor type O |
| chr15_+_45023137 | 0.01 |
ENST00000674211.1
ENST00000267814.14 |
SORD
|
sorbitol dehydrogenase |
| chr17_+_7884783 | 0.01 |
ENST00000380358.9
|
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr2_+_203706475 | 0.00 |
ENST00000374481.7
ENST00000458610.6 |
CD28
|
CD28 molecule |
| chr1_+_218345326 | 0.00 |
ENST00000366930.9
|
TGFB2
|
transforming growth factor beta 2 |
| chr6_+_26045374 | 0.00 |
ENST00000612966.3
|
H3C3
|
H3 clustered histone 3 |
| chr5_+_145937793 | 0.00 |
ENST00000511217.1
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr20_+_59721210 | 0.00 |
ENST00000395636.6
ENST00000361300.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.2 | 0.7 | GO:2001226 | negative regulation of anion channel activity(GO:0010360) negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.2 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.0 | 0.1 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:1901874 | negative regulation of post-translational protein modification(GO:1901874) |
| 0.0 | 0.2 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.3 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.1 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.2 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |