Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for PRRX1_ALX4_PHOX2A
Z-value: 0.43
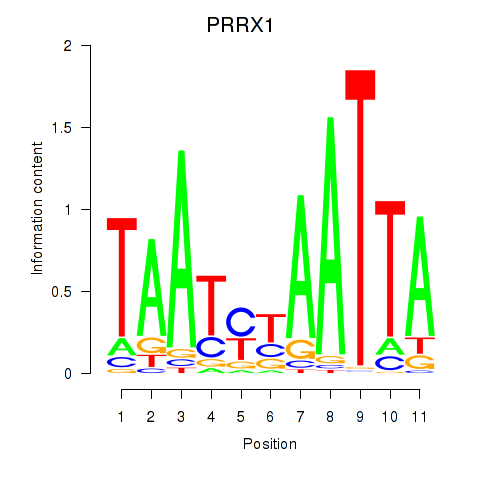
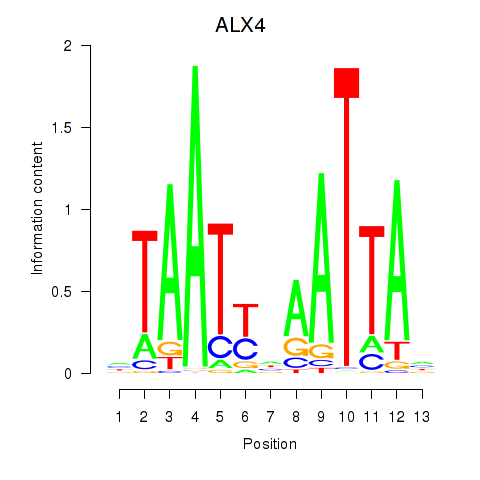
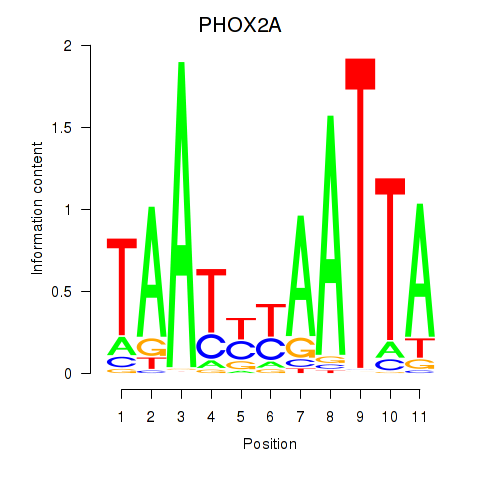
Transcription factors associated with PRRX1_ALX4_PHOX2A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRRX1
|
ENSG00000116132.12 | PRRX1 |
|
ALX4
|
ENSG00000052850.8 | ALX4 |
|
PHOX2A
|
ENSG00000165462.5 | PHOX2A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PRRX1 | hg38_v1_chr1_+_170664121_170664144 | -0.35 | 8.3e-02 | Click! |
| ALX4 | hg38_v1_chr11_-_44310126_44310152 | 0.30 | 1.5e-01 | Click! |
| PHOX2A | hg38_v1_chr11_-_72241192_72241275, hg38_v1_chr11_-_72244166_72244184 | -0.06 | 7.9e-01 | Click! |
Activity profile of PRRX1_ALX4_PHOX2A motif
Sorted Z-values of PRRX1_ALX4_PHOX2A motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PRRX1_ALX4_PHOX2A
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_91906280 | 1.83 |
ENST00000370399.6
|
TGFBR3
|
transforming growth factor beta receptor 3 |
| chr1_+_84181630 | 1.23 |
ENST00000610457.1
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr2_+_178320228 | 0.82 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein like 6 |
| chr6_-_75363003 | 0.78 |
ENST00000370020.1
|
FILIP1
|
filamin A interacting protein 1 |
| chr3_-_27722699 | 0.68 |
ENST00000461503.2
|
EOMES
|
eomesodermin |
| chr9_-_123184233 | 0.60 |
ENST00000447404.6
|
STRBP
|
spermatid perinuclear RNA binding protein |
| chr3_-_125055987 | 0.52 |
ENST00000311127.9
|
HEG1
|
heart development protein with EGF like domains 1 |
| chr18_+_616711 | 0.51 |
ENST00000579494.1
|
CLUL1
|
clusterin like 1 |
| chr18_+_616672 | 0.50 |
ENST00000338387.11
|
CLUL1
|
clusterin like 1 |
| chr10_+_68106109 | 0.47 |
ENST00000540630.5
ENST00000354393.6 |
MYPN
|
myopalladin |
| chr3_-_27722316 | 0.47 |
ENST00000449599.4
|
EOMES
|
eomesodermin |
| chr1_+_84164962 | 0.47 |
ENST00000614872.4
ENST00000394839.6 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr5_-_111976925 | 0.46 |
ENST00000395634.7
|
NREP
|
neuronal regeneration related protein |
| chr3_-_18438767 | 0.45 |
ENST00000454909.6
|
SATB1
|
SATB homeobox 1 |
| chr5_-_111758061 | 0.45 |
ENST00000509979.5
ENST00000513100.5 ENST00000508161.5 ENST00000455559.6 |
NREP
|
neuronal regeneration related protein |
| chr2_-_218010202 | 0.44 |
ENST00000646520.1
|
TNS1
|
tensin 1 |
| chr1_-_93681829 | 0.43 |
ENST00000260502.11
|
BCAR3
|
BCAR3 adaptor protein, NSP family member |
| chr8_-_92095215 | 0.42 |
ENST00000360348.6
ENST00000520428.5 ENST00000518992.5 ENST00000520556.5 ENST00000518317.5 ENST00000521319.5 ENST00000521375.5 ENST00000518449.5 ENST00000613886.4 |
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr3_-_142029108 | 0.41 |
ENST00000497579.5
|
TFDP2
|
transcription factor Dp-2 |
| chr10_+_35175586 | 0.40 |
ENST00000494479.5
ENST00000463314.5 ENST00000342105.7 ENST00000495301.1 |
CREM
|
cAMP responsive element modulator |
| chr6_-_132659178 | 0.39 |
ENST00000275216.3
|
TAAR1
|
trace amine associated receptor 1 |
| chr2_-_55334529 | 0.39 |
ENST00000645860.1
ENST00000642563.1 ENST00000647396.1 |
CCDC88A
|
coiled-coil domain containing 88A |
| chr6_+_116369837 | 0.38 |
ENST00000645988.1
|
DSE
|
dermatan sulfate epimerase |
| chr17_-_40994159 | 0.34 |
ENST00000391586.3
|
KRTAP3-3
|
keratin associated protein 3-3 |
| chr9_+_133636355 | 0.34 |
ENST00000393056.8
|
DBH
|
dopamine beta-hydroxylase |
| chr2_-_55269038 | 0.29 |
ENST00000417363.5
ENST00000412530.1 ENST00000366137.6 ENST00000420637.5 |
MTIF2
|
mitochondrial translational initiation factor 2 |
| chr12_+_19205294 | 0.29 |
ENST00000424268.5
|
PLEKHA5
|
pleckstrin homology domain containing A5 |
| chr1_-_21050952 | 0.28 |
ENST00000264211.12
|
EIF4G3
|
eukaryotic translation initiation factor 4 gamma 3 |
| chr3_+_98149326 | 0.27 |
ENST00000437310.1
|
OR5H14
|
olfactory receptor family 5 subfamily H member 14 |
| chr6_+_30914205 | 0.26 |
ENST00000672801.1
ENST00000321897.9 ENST00000625423.2 ENST00000676266.1 ENST00000428017.5 |
VARS2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr18_+_44700796 | 0.26 |
ENST00000677130.1
|
SETBP1
|
SET binding protein 1 |
| chr6_+_30914329 | 0.25 |
ENST00000541562.6
|
VARS2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr9_+_128566741 | 0.25 |
ENST00000630866.1
|
SPTAN1
|
spectrin alpha, non-erythrocytic 1 |
| chr2_-_182427014 | 0.24 |
ENST00000409365.5
ENST00000351439.9 |
PDE1A
|
phosphodiesterase 1A |
| chr12_-_7503744 | 0.24 |
ENST00000396620.7
ENST00000432237.3 |
CD163
|
CD163 molecule |
| chr12_-_91153149 | 0.24 |
ENST00000550758.1
|
DCN
|
decorin |
| chr20_+_36091409 | 0.23 |
ENST00000202028.9
|
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr3_-_151316795 | 0.23 |
ENST00000260843.5
|
GPR87
|
G protein-coupled receptor 87 |
| chr14_+_22516273 | 0.23 |
ENST00000390510.1
|
TRAJ27
|
T cell receptor alpha joining 27 |
| chr2_-_201739205 | 0.22 |
ENST00000681558.1
ENST00000681495.1 |
ALS2
|
alsin Rho guanine nucleotide exchange factor ALS2 |
| chr6_+_121437378 | 0.21 |
ENST00000650427.1
ENST00000647564.1 |
GJA1
|
gap junction protein alpha 1 |
| chr5_-_67196791 | 0.21 |
ENST00000256447.5
|
CD180
|
CD180 molecule |
| chr2_+_189857393 | 0.21 |
ENST00000452382.1
|
PMS1
|
PMS1 homolog 1, mismatch repair system component |
| chr5_+_36606355 | 0.21 |
ENST00000681909.1
ENST00000513903.5 ENST00000681795.1 ENST00000680125.1 ENST00000612708.5 ENST00000680232.1 ENST00000681776.1 ENST00000681926.1 ENST00000679958.1 ENST00000265113.9 ENST00000504121.5 ENST00000512374.1 ENST00000613445.5 ENST00000679983.1 |
SLC1A3
|
solute carrier family 1 member 3 |
| chr9_-_14314519 | 0.21 |
ENST00000397581.6
|
NFIB
|
nuclear factor I B |
| chrX_+_77910656 | 0.21 |
ENST00000343533.9
ENST00000341514.11 ENST00000645454.1 ENST00000642651.1 ENST00000644362.1 |
ATP7A
PGK1
|
ATPase copper transporting alpha phosphoglycerate kinase 1 |
| chr21_-_34526815 | 0.20 |
ENST00000492600.1
|
RCAN1
|
regulator of calcineurin 1 |
| chr9_-_14314567 | 0.20 |
ENST00000397579.6
|
NFIB
|
nuclear factor I B |
| chr12_-_106987068 | 0.20 |
ENST00000548101.1
ENST00000550496.1 ENST00000552029.1 |
MTERF2
|
mitochondrial transcription termination factor 2 |
| chr5_-_55173173 | 0.20 |
ENST00000296733.5
ENST00000322374.10 ENST00000381375.7 |
CDC20B
|
cell division cycle 20B |
| chr4_-_142305826 | 0.19 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr12_-_7503841 | 0.18 |
ENST00000359156.8
|
CD163
|
CD163 molecule |
| chr12_+_20815672 | 0.18 |
ENST00000261196.6
ENST00000381541.7 ENST00000540229.1 |
SLCO1B3
SLCO1B3-SLCO1B7
|
solute carrier organic anion transporter family member 1B3 SLCO1B3-SLCO1B7 readthrough |
| chr14_-_37595224 | 0.18 |
ENST00000250448.5
|
FOXA1
|
forkhead box A1 |
| chr20_+_33235987 | 0.18 |
ENST00000375422.6
ENST00000375413.8 ENST00000354297.9 |
BPIFA1
|
BPI fold containing family A member 1 |
| chr11_+_66509079 | 0.18 |
ENST00000419755.3
|
ENSG00000256349.1
|
novel protein |
| chr9_+_2159672 | 0.18 |
ENST00000634343.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr11_-_40294089 | 0.17 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr15_+_57219411 | 0.17 |
ENST00000543579.5
ENST00000537840.5 ENST00000343827.7 |
TCF12
|
transcription factor 12 |
| chr12_-_106987131 | 0.17 |
ENST00000240050.9
ENST00000392830.6 |
MTERF2
|
mitochondrial transcription termination factor 2 |
| chr7_+_148590760 | 0.17 |
ENST00000307003.3
|
C7orf33
|
chromosome 7 open reading frame 33 |
| chr12_-_91179472 | 0.17 |
ENST00000550099.5
ENST00000546391.5 |
DCN
|
decorin |
| chr6_-_41071825 | 0.17 |
ENST00000468811.5
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr15_+_75336018 | 0.17 |
ENST00000567195.5
|
COMMD4
|
COMM domain containing 4 |
| chr16_+_68085344 | 0.16 |
ENST00000575270.5
ENST00000346183.8 |
NFATC3
|
nuclear factor of activated T cells 3 |
| chr21_-_34526850 | 0.16 |
ENST00000481448.5
ENST00000381132.6 |
RCAN1
|
regulator of calcineurin 1 |
| chr6_+_27824084 | 0.16 |
ENST00000355057.3
|
H4C11
|
H4 clustered histone 11 |
| chr4_-_99435396 | 0.16 |
ENST00000209665.8
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr11_+_108116688 | 0.15 |
ENST00000672284.1
|
ACAT1
|
acetyl-CoA acetyltransferase 1 |
| chr1_-_165445220 | 0.15 |
ENST00000619224.1
|
RXRG
|
retinoid X receptor gamma |
| chr13_+_53028806 | 0.15 |
ENST00000219022.3
|
OLFM4
|
olfactomedin 4 |
| chr18_-_55321986 | 0.14 |
ENST00000570287.6
|
TCF4
|
transcription factor 4 |
| chr2_-_58241259 | 0.14 |
ENST00000481670.2
ENST00000403676.5 ENST00000427708.6 ENST00000403295.7 ENST00000233741.9 ENST00000446381.5 ENST00000417361.1 ENST00000402135.7 ENST00000449070.5 |
FANCL
|
FA complementation group L |
| chr8_-_6563409 | 0.14 |
ENST00000325203.9
|
ANGPT2
|
angiopoietin 2 |
| chr16_+_68085420 | 0.14 |
ENST00000349223.9
|
NFATC3
|
nuclear factor of activated T cells 3 |
| chr1_-_68232514 | 0.14 |
ENST00000262348.9
ENST00000370973.2 ENST00000370971.1 |
WLS
|
Wnt ligand secretion mediator |
| chr19_-_7021431 | 0.14 |
ENST00000636986.2
ENST00000637800.1 |
MBD3L2B
|
methyl-CpG binding domain protein 3 like 2B |
| chrX_-_19799751 | 0.14 |
ENST00000379698.8
|
SH3KBP1
|
SH3 domain containing kinase binding protein 1 |
| chr1_-_165445088 | 0.14 |
ENST00000359842.10
|
RXRG
|
retinoid X receptor gamma |
| chr9_+_2159850 | 0.14 |
ENST00000416751.2
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr12_-_23584600 | 0.14 |
ENST00000396007.6
|
SOX5
|
SRY-box transcription factor 5 |
| chr6_+_26087417 | 0.14 |
ENST00000357618.10
ENST00000309234.10 |
HFE
|
homeostatic iron regulator |
| chr20_+_16748358 | 0.13 |
ENST00000246081.3
|
OTOR
|
otoraplin |
| chr13_+_57140918 | 0.13 |
ENST00000377931.1
ENST00000614894.4 |
PRR20A
PRR20C
|
proline rich 20A proline rich 20C |
| chr18_-_55322215 | 0.13 |
ENST00000457482.7
|
TCF4
|
transcription factor 4 |
| chr15_-_52678560 | 0.13 |
ENST00000562351.2
ENST00000261844.11 ENST00000399202.8 ENST00000562135.5 |
FAM214A
|
family with sequence similarity 214 member A |
| chr19_-_11346228 | 0.13 |
ENST00000588560.5
ENST00000592952.5 |
TMEM205
|
transmembrane protein 205 |
| chr22_-_28306645 | 0.13 |
ENST00000612946.4
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr8_-_85341659 | 0.13 |
ENST00000522389.5
|
CA1
|
carbonic anhydrase 1 |
| chr4_+_185204237 | 0.12 |
ENST00000618785.4
ENST00000504273.5 |
SNX25
|
sorting nexin 25 |
| chr16_+_68085552 | 0.12 |
ENST00000329524.8
|
NFATC3
|
nuclear factor of activated T cells 3 |
| chr6_+_132552693 | 0.12 |
ENST00000275200.1
|
TAAR8
|
trace amine associated receptor 8 |
| chr19_-_11346259 | 0.12 |
ENST00000590788.1
ENST00000354882.10 ENST00000586590.5 ENST00000589555.5 ENST00000586956.5 ENST00000593256.6 ENST00000447337.5 ENST00000591677.5 ENST00000586701.1 ENST00000589655.1 |
TMEM205
RAB3D
|
transmembrane protein 205 RAB3D, member RAS oncogene family |
| chr17_-_48613468 | 0.12 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr1_-_68232539 | 0.12 |
ENST00000370976.7
ENST00000354777.6 |
WLS
|
Wnt ligand secretion mediator |
| chr19_+_49513353 | 0.12 |
ENST00000596975.5
|
FCGRT
|
Fc fragment of IgG receptor and transporter |
| chrX_+_37780049 | 0.12 |
ENST00000378588.5
|
CYBB
|
cytochrome b-245 beta chain |
| chr16_-_67483541 | 0.11 |
ENST00000290953.3
|
AGRP
|
agouti related neuropeptide |
| chr16_-_71577082 | 0.11 |
ENST00000355962.5
|
TAT
|
tyrosine aminotransferase |
| chr1_+_15341744 | 0.11 |
ENST00000444385.5
|
FHAD1
|
forkhead associated phosphopeptide binding domain 1 |
| chr7_+_107583919 | 0.11 |
ENST00000491150.5
|
BCAP29
|
B cell receptor associated protein 29 |
| chr5_-_175961324 | 0.11 |
ENST00000432305.6
ENST00000505969.1 |
THOC3
|
THO complex 3 |
| chr2_-_208129824 | 0.11 |
ENST00000282141.4
|
CRYGC
|
crystallin gamma C |
| chr7_-_111392915 | 0.11 |
ENST00000450877.5
|
IMMP2L
|
inner mitochondrial membrane peptidase subunit 2 |
| chr17_-_9791586 | 0.11 |
ENST00000571134.2
|
DHRS7C
|
dehydrogenase/reductase 7C |
| chr14_-_21098848 | 0.10 |
ENST00000556174.5
ENST00000554478.5 ENST00000553980.1 ENST00000421093.6 |
ZNF219
|
zinc finger protein 219 |
| chr9_-_92536031 | 0.10 |
ENST00000344604.9
ENST00000375540.5 |
ECM2
|
extracellular matrix protein 2 |
| chr13_-_52406160 | 0.10 |
ENST00000258613.5
|
THSD1
|
thrombospondin type 1 domain containing 1 |
| chr4_-_142305935 | 0.10 |
ENST00000511838.5
|
INPP4B
|
inositol polyphosphate-4-phosphatase type II B |
| chr4_-_99435336 | 0.10 |
ENST00000437033.7
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr8_+_26293112 | 0.10 |
ENST00000523925.5
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2 regulatory subunit Balpha |
| chr5_+_141192330 | 0.10 |
ENST00000239446.6
|
PCDHB10
|
protocadherin beta 10 |
| chr12_-_81598332 | 0.10 |
ENST00000443686.7
|
PPFIA2
|
PTPRF interacting protein alpha 2 |
| chr9_+_72577369 | 0.10 |
ENST00000651183.1
|
TMC1
|
transmembrane channel like 1 |
| chr10_+_48684859 | 0.10 |
ENST00000360890.6
ENST00000325239.11 |
WDFY4
|
WDFY family member 4 |
| chr12_+_12725897 | 0.09 |
ENST00000326765.10
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr3_-_149576203 | 0.09 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr15_+_51377247 | 0.09 |
ENST00000396399.6
|
GLDN
|
gliomedin |
| chr1_+_180632001 | 0.09 |
ENST00000367590.9
ENST00000367589.3 |
XPR1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr2_+_162318884 | 0.09 |
ENST00000446271.5
ENST00000429691.6 |
GCA
|
grancalcin |
| chr2_+_176129680 | 0.09 |
ENST00000429017.2
ENST00000313173.6 |
HOXD8
|
homeobox D8 |
| chr5_+_36151989 | 0.09 |
ENST00000274254.9
|
SKP2
|
S-phase kinase associated protein 2 |
| chr12_+_133181409 | 0.09 |
ENST00000416488.5
ENST00000228289.9 ENST00000541211.6 ENST00000536435.7 ENST00000500625.7 ENST00000539248.6 ENST00000542711.6 ENST00000536899.6 ENST00000542986.6 ENST00000611984.4 ENST00000541975.2 |
ZNF268
|
zinc finger protein 268 |
| chr6_+_42929127 | 0.09 |
ENST00000394142.7
|
CNPY3
|
canopy FGF signaling regulator 3 |
| chr15_+_75336053 | 0.09 |
ENST00000564815.5
ENST00000338995.10 ENST00000267935.13 |
COMMD4
|
COMM domain containing 4 |
| chr7_+_23299306 | 0.08 |
ENST00000466681.2
|
MALSU1
|
mitochondrial assembly of ribosomal large subunit 1 |
| chr12_-_10802627 | 0.08 |
ENST00000240687.2
|
TAS2R7
|
taste 2 receptor member 7 |
| chr9_+_34652167 | 0.08 |
ENST00000441545.7
ENST00000553620.5 |
IL11RA
|
interleukin 11 receptor subunit alpha |
| chr17_+_3197519 | 0.08 |
ENST00000381951.1
|
OR1A2
|
olfactory receptor family 1 subfamily A member 2 |
| chr21_+_46635595 | 0.08 |
ENST00000451211.6
ENST00000458387.6 ENST00000397638.6 ENST00000291705.11 ENST00000397637.5 ENST00000334494.8 ENST00000397628.5 ENST00000355680.8 ENST00000440086.5 |
PRMT2
|
protein arginine methyltransferase 2 |
| chr7_+_90709530 | 0.08 |
ENST00000406263.5
|
CDK14
|
cyclin dependent kinase 14 |
| chr3_-_157160751 | 0.08 |
ENST00000461804.5
|
CCNL1
|
cyclin L1 |
| chr17_+_46511511 | 0.08 |
ENST00000576629.5
|
LRRC37A2
|
leucine rich repeat containing 37 member A2 |
| chr1_+_244051275 | 0.08 |
ENST00000358704.4
|
ZBTB18
|
zinc finger and BTB domain containing 18 |
| chr15_+_75336078 | 0.08 |
ENST00000567377.5
ENST00000562789.5 ENST00000568301.1 |
COMMD4
|
COMM domain containing 4 |
| chr5_-_9630351 | 0.08 |
ENST00000382492.4
|
TAS2R1
|
taste 2 receptor member 1 |
| chr2_+_165294031 | 0.08 |
ENST00000283256.10
|
SCN2A
|
sodium voltage-gated channel alpha subunit 2 |
| chr16_+_21233672 | 0.08 |
ENST00000311620.7
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr12_+_57610150 | 0.08 |
ENST00000333972.11
|
ARHGEF25
|
Rho guanine nucleotide exchange factor 25 |
| chr12_-_89352395 | 0.08 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr12_-_68225806 | 0.07 |
ENST00000229134.5
|
IL26
|
interleukin 26 |
| chr12_-_91004965 | 0.07 |
ENST00000261172.8
|
EPYC
|
epiphycan |
| chr19_-_57974527 | 0.07 |
ENST00000314391.3
|
C19orf18
|
chromosome 19 open reading frame 18 |
| chr15_-_23687290 | 0.07 |
ENST00000649030.2
|
NDN
|
necdin, MAGE family member |
| chr8_-_17895403 | 0.07 |
ENST00000381840.5
ENST00000398054.5 |
FGL1
|
fibrinogen like 1 |
| chr1_-_151175966 | 0.07 |
ENST00000441701.1
ENST00000295314.9 |
TMOD4
|
tropomodulin 4 |
| chr8_-_17895487 | 0.07 |
ENST00000427924.5
ENST00000381841.4 |
FGL1
|
fibrinogen like 1 |
| chr16_+_86566821 | 0.07 |
ENST00000649859.1
|
FOXC2
|
forkhead box C2 |
| chr1_+_67166448 | 0.07 |
ENST00000347310.10
|
IL23R
|
interleukin 23 receptor |
| chr18_-_24311495 | 0.07 |
ENST00000357041.8
|
OSBPL1A
|
oxysterol binding protein like 1A |
| chr12_+_20810698 | 0.07 |
ENST00000540853.5
ENST00000381545.8 |
SLCO1B3
|
solute carrier organic anion transporter family member 1B3 |
| chr5_+_141392616 | 0.07 |
ENST00000398604.3
|
PCDHGA8
|
protocadherin gamma subfamily A, 8 |
| chr5_+_36152077 | 0.06 |
ENST00000546211.6
ENST00000620197.5 ENST00000678270.1 ENST00000679015.1 ENST00000678580.1 ENST00000274255.11 ENST00000508514.5 |
SKP2
|
S-phase kinase associated protein 2 |
| chrX_+_35919725 | 0.06 |
ENST00000297866.9
ENST00000378653.8 |
CFAP47
|
cilia and flagella associated protein 47 |
| chr5_+_42548043 | 0.06 |
ENST00000618088.4
ENST00000612382.4 |
GHR
|
growth hormone receptor |
| chr1_+_24556087 | 0.06 |
ENST00000374392.3
|
NCMAP
|
non-compact myelin associated protein |
| chr12_+_53461015 | 0.06 |
ENST00000553064.6
ENST00000547859.2 |
PCBP2
|
poly(rC) binding protein 2 |
| chr11_-_95232514 | 0.06 |
ENST00000634898.1
ENST00000542176.1 ENST00000278499.6 |
SESN3
|
sestrin 3 |
| chr5_+_141135199 | 0.06 |
ENST00000231134.8
ENST00000623915.1 |
PCDHB5
|
protocadherin beta 5 |
| chr1_+_158845798 | 0.06 |
ENST00000438394.1
|
MNDA
|
myeloid cell nuclear differentiation antigen |
| chr21_-_42395943 | 0.06 |
ENST00000398405.5
|
TMPRSS3
|
transmembrane serine protease 3 |
| chr12_-_89352487 | 0.06 |
ENST00000548755.1
ENST00000279488.8 |
DUSP6
|
dual specificity phosphatase 6 |
| chr12_+_59689337 | 0.06 |
ENST00000261187.8
|
SLC16A7
|
solute carrier family 16 member 7 |
| chr10_-_29634964 | 0.06 |
ENST00000375398.6
ENST00000355867.8 |
SVIL
|
supervillin |
| chr11_-_82997394 | 0.06 |
ENST00000525117.5
ENST00000532548.5 |
RAB30
|
RAB30, member RAS oncogene family |
| chr19_+_7030578 | 0.06 |
ENST00000329753.5
|
MBD3L5
|
methyl-CpG binding domain protein 3 like 5 |
| chr19_+_55769118 | 0.06 |
ENST00000341750.5
|
RFPL4AL1
|
ret finger protein like 4A like 1 |
| chr12_-_9999176 | 0.06 |
ENST00000298527.10
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1 member B |
| chr3_+_69936629 | 0.06 |
ENST00000394348.2
ENST00000531774.1 |
MITF
|
melanocyte inducing transcription factor |
| chr3_-_197573323 | 0.06 |
ENST00000358186.6
ENST00000431056.5 |
BDH1
|
3-hydroxybutyrate dehydrogenase 1 |
| chr19_-_6393205 | 0.05 |
ENST00000595047.5
|
GTF2F1
|
general transcription factor IIF subunit 1 |
| chr12_-_124567464 | 0.05 |
ENST00000458234.5
|
NCOR2
|
nuclear receptor corepressor 2 |
| chr19_+_13731744 | 0.05 |
ENST00000586600.5
|
CCDC130
|
coiled-coil domain containing 130 |
| chr12_-_118190510 | 0.05 |
ENST00000540561.5
ENST00000537952.1 ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr6_+_44219595 | 0.05 |
ENST00000393844.7
ENST00000652680.1 ENST00000643028.2 ENST00000371713.6 |
SLC29A1
|
solute carrier family 29 member 1 (Augustine blood group) |
| chr7_+_136868622 | 0.05 |
ENST00000680005.1
ENST00000445907.6 |
CHRM2
|
cholinergic receptor muscarinic 2 |
| chr15_+_45252228 | 0.05 |
ENST00000560438.5
ENST00000347644.8 |
SLC28A2
|
solute carrier family 28 member 2 |
| chr7_+_90709231 | 0.05 |
ENST00000446790.5
ENST00000265741.7 |
CDK14
|
cyclin dependent kinase 14 |
| chr15_-_65115185 | 0.05 |
ENST00000559089.6
|
UBAP1L
|
ubiquitin associated protein 1 like |
| chr6_+_44219527 | 0.05 |
ENST00000651428.1
|
SLC29A1
|
solute carrier family 29 member 1 (Augustine blood group) |
| chr10_-_103351074 | 0.05 |
ENST00000337211.8
|
PCGF6
|
polycomb group ring finger 6 |
| chr3_+_138621225 | 0.05 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr19_-_11346196 | 0.05 |
ENST00000586218.5
|
TMEM205
|
transmembrane protein 205 |
| chr7_-_93890744 | 0.05 |
ENST00000650573.1
ENST00000222543.11 ENST00000649913.1 ENST00000647793.1 |
TFPI2
|
tissue factor pathway inhibitor 2 |
| chr6_+_31927486 | 0.05 |
ENST00000442278.6
|
C2
|
complement C2 |
| chr10_-_103351133 | 0.05 |
ENST00000369847.4
|
PCGF6
|
polycomb group ring finger 6 |
| chr12_-_10807286 | 0.04 |
ENST00000240615.3
|
TAS2R8
|
taste 2 receptor member 8 |
| chr19_-_51417581 | 0.04 |
ENST00000442846.7
ENST00000530476.1 |
SIGLEC10
|
sialic acid binding Ig like lectin 10 |
| chr1_-_248277976 | 0.04 |
ENST00000641220.1
|
OR2T33
|
olfactory receptor family 2 subfamily T member 33 |
| chr1_+_151766655 | 0.04 |
ENST00000400999.7
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr5_+_141382702 | 0.04 |
ENST00000617050.1
ENST00000518325.2 |
PCDHGA7
|
protocadherin gamma subfamily A, 7 |
| chr13_-_94479671 | 0.04 |
ENST00000377028.10
ENST00000446125.1 |
DCT
|
dopachrome tautomerase |
| chr5_+_90640718 | 0.04 |
ENST00000640403.1
|
ADGRV1
|
adhesion G protein-coupled receptor V1 |
| chr7_+_142384328 | 0.04 |
ENST00000390361.3
|
TRBV7-3
|
T cell receptor beta variable 7-3 |
| chr16_+_51553436 | 0.04 |
ENST00000565308.2
|
HNRNPA1P48
|
heterogeneous nuclear ribonucleoprotein A1 pseudogene 48 |
| chr15_+_48191648 | 0.04 |
ENST00000646012.1
ENST00000561127.5 ENST00000647546.1 ENST00000559641.5 ENST00000417307.3 |
SLC12A1
CTXN2
|
solute carrier family 12 member 1 cortexin 2 |
| chr15_-_37101205 | 0.04 |
ENST00000338564.9
ENST00000558313.5 ENST00000340545.9 |
MEIS2
|
Meis homeobox 2 |
| chr17_-_10373002 | 0.04 |
ENST00000252172.9
ENST00000418404.8 |
MYH13
|
myosin heavy chain 13 |
| chr5_-_135399863 | 0.04 |
ENST00000510038.1
ENST00000304332.8 |
MACROH2A1
|
macroH2A.1 histone |
| chr17_+_76467597 | 0.04 |
ENST00000392492.8
|
AANAT
|
aralkylamine N-acetyltransferase |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.8 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.4 | 1.2 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.2 | 1.7 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.2 | 0.5 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.4 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 0.1 | 0.3 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.1 | 0.2 | GO:0071284 | copper ion export(GO:0060003) cellular response to lead ion(GO:0071284) |
| 0.1 | 0.3 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.4 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.2 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.1 | 0.2 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.1 | 0.2 | GO:0072019 | proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.0 | 0.3 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.0 | 0.1 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.0 | 0.2 | GO:0042710 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.0 | 0.1 | GO:1904432 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.0 | 0.2 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.1 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.4 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.5 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.4 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.2 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.5 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.7 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.2 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.5 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.2 | GO:0009449 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.1 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.9 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.6 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.0 | GO:0015847 | putrescine transport(GO:0015847) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.0 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.0 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.0 | GO:0034414 | tRNA 3'-trailer cleavage, endonucleolytic(GO:0034414) tRNA 3'-trailer cleavage(GO:0042779) mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.0 | 0.2 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.0 | GO:1903259 | exon-exon junction complex disassembly(GO:1903259) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 1.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.8 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.3 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.3 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.2 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 1.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.4 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 0.5 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.3 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.2 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) phosphoglycerate kinase activity(GO:0004618) copper-transporting ATPase activity(GO:0043682) |
| 0.1 | 0.5 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.1 | 0.3 | GO:0052828 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 0.3 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.2 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.2 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.4 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.0 | GO:0015489 | polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 0.0 | 1.1 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.7 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.0 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.0 | 0.1 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.0 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.0 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 0.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.0 | GO:1902271 | D3 vitamins binding(GO:1902271) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.2 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.3 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |