Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
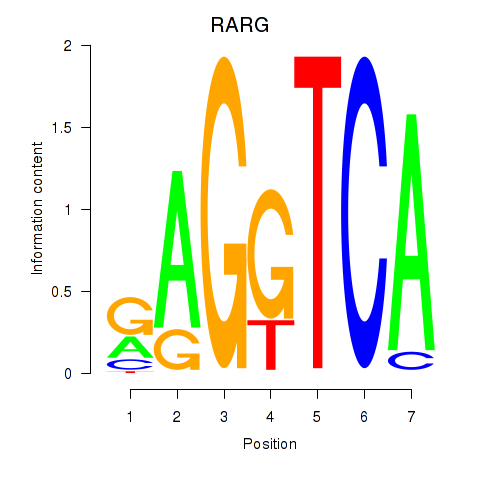
Results for RARG
Z-value: 0.77
Transcription factors associated with RARG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RARG
|
ENSG00000172819.17 | RARG |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RARG | hg38_v1_chr12_-_53220229_53220272, hg38_v1_chr12_-_53220377_53220413 | 0.39 | 5.3e-02 | Click! |
Activity profile of RARG motif
Sorted Z-values of RARG motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RARG
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_132570322 | 0.90 |
ENST00000275198.1
|
TAAR6
|
trace amine associated receptor 6 |
| chr19_+_50415799 | 0.70 |
ENST00000599632.1
|
ENSG00000142539.9
|
novel protein |
| chr8_+_62248591 | 0.64 |
ENST00000519049.6
|
NKAIN3
|
sodium/potassium transporting ATPase interacting 3 |
| chr1_+_231162052 | 0.60 |
ENST00000366653.6
ENST00000444294.7 |
TRIM67
|
tripartite motif containing 67 |
| chr1_+_222928415 | 0.59 |
ENST00000284476.7
|
DISP1
|
dispatched RND transporter family member 1 |
| chr5_-_151093566 | 0.53 |
ENST00000521001.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr8_+_66043413 | 0.53 |
ENST00000522619.1
|
DNAJC5B
|
DnaJ heat shock protein family (Hsp40) member C5 beta |
| chr12_+_52233235 | 0.53 |
ENST00000331817.6
|
KRT7
|
keratin 7 |
| chr3_-_116444983 | 0.52 |
ENST00000333617.8
|
LSAMP
|
limbic system associated membrane protein |
| chr1_-_54623518 | 0.51 |
ENST00000302250.7
ENST00000371304.2 |
FAM151A
|
family with sequence similarity 151 member A |
| chr1_+_222815065 | 0.51 |
ENST00000675039.1
ENST00000675961.1 |
DISP1
|
dispatched RND transporter family member 1 |
| chr2_+_218607914 | 0.48 |
ENST00000417849.5
|
PLCD4
|
phospholipase C delta 4 |
| chr12_-_91182652 | 0.47 |
ENST00000552145.5
ENST00000546745.5 |
DCN
|
decorin |
| chr20_-_62475983 | 0.47 |
ENST00000252997.3
|
GATA5
|
GATA binding protein 5 |
| chr1_+_32362537 | 0.46 |
ENST00000373534.4
|
TSSK3
|
testis specific serine kinase 3 |
| chr14_-_76826229 | 0.45 |
ENST00000557497.1
|
ANGEL1
|
angel homolog 1 |
| chr1_-_19923617 | 0.44 |
ENST00000375116.3
|
PLA2G2E
|
phospholipase A2 group IIE |
| chr1_-_173917281 | 0.44 |
ENST00000367698.4
|
SERPINC1
|
serpin family C member 1 |
| chr7_-_138679045 | 0.43 |
ENST00000419765.4
|
SVOPL
|
SVOP like |
| chr2_+_74513441 | 0.43 |
ENST00000621092.1
|
TLX2
|
T cell leukemia homeobox 2 |
| chr19_+_11798514 | 0.42 |
ENST00000323169.10
ENST00000450087.1 |
ZNF491
|
zinc finger protein 491 |
| chr19_-_51065067 | 0.42 |
ENST00000595547.5
ENST00000335422.3 ENST00000595793.6 ENST00000596955.1 |
KLK13
|
kallikrein related peptidase 13 |
| chr3_-_126517764 | 0.41 |
ENST00000290868.7
|
UROC1
|
urocanate hydratase 1 |
| chr4_-_48080172 | 0.41 |
ENST00000507351.1
|
TXK
|
TXK tyrosine kinase |
| chr7_-_8262668 | 0.40 |
ENST00000446305.1
|
ICA1
|
islet cell autoantigen 1 |
| chr6_-_41071825 | 0.40 |
ENST00000468811.5
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr13_-_33185994 | 0.40 |
ENST00000255486.8
|
STARD13
|
StAR related lipid transfer domain containing 13 |
| chr6_-_119078642 | 0.40 |
ENST00000621231.4
ENST00000338891.12 |
FAM184A
|
family with sequence similarity 184 member A |
| chr19_-_40690553 | 0.40 |
ENST00000598779.5
|
NUMBL
|
NUMB like endocytic adaptor protein |
| chr16_-_88686453 | 0.39 |
ENST00000332281.6
|
SNAI3
|
snail family transcriptional repressor 3 |
| chr16_-_50681328 | 0.39 |
ENST00000300590.7
|
SNX20
|
sorting nexin 20 |
| chr8_+_12951583 | 0.38 |
ENST00000528753.2
|
TRMT9B
|
tRNA methyltransferase 9B (putative) |
| chr12_-_7128873 | 0.38 |
ENST00000542370.1
ENST00000266560.8 |
RBP5
|
retinol binding protein 5 |
| chr8_+_12951325 | 0.38 |
ENST00000400069.7
|
TRMT9B
|
tRNA methyltransferase 9B (putative) |
| chr12_+_12785652 | 0.37 |
ENST00000356591.5
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr9_+_117704168 | 0.37 |
ENST00000472304.2
ENST00000394487.5 |
TLR4
|
toll like receptor 4 |
| chr7_-_27143672 | 0.36 |
ENST00000222726.4
|
HOXA5
|
homeobox A5 |
| chr9_-_34381578 | 0.35 |
ENST00000379133.7
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr1_+_160127672 | 0.34 |
ENST00000447527.1
|
ATP1A2
|
ATPase Na+/K+ transporting subunit alpha 2 |
| chr1_+_222815021 | 0.34 |
ENST00000675850.1
ENST00000495684.2 |
DISP1
|
dispatched RND transporter family member 1 |
| chr12_-_52473798 | 0.34 |
ENST00000252250.7
|
KRT6C
|
keratin 6C |
| chr11_+_64237420 | 0.34 |
ENST00000541681.1
|
VEGFB
|
vascular endothelial growth factor B |
| chr19_+_6464491 | 0.33 |
ENST00000308243.7
|
CRB3
|
crumbs cell polarity complex component 3 |
| chr1_-_152159227 | 0.33 |
ENST00000316073.3
|
RPTN
|
repetin |
| chr20_+_2814981 | 0.33 |
ENST00000603872.2
ENST00000380589.4 |
C20orf141
|
chromosome 20 open reading frame 141 |
| chr5_+_150166770 | 0.33 |
ENST00000231656.13
|
CDX1
|
caudal type homeobox 1 |
| chr18_-_48950960 | 0.33 |
ENST00000262158.8
|
SMAD7
|
SMAD family member 7 |
| chr8_-_70071226 | 0.33 |
ENST00000276594.3
|
PRDM14
|
PR/SET domain 14 |
| chr2_-_31414694 | 0.32 |
ENST00000379416.4
|
XDH
|
xanthine dehydrogenase |
| chr7_-_150341615 | 0.32 |
ENST00000223271.8
ENST00000466675.5 ENST00000482669.1 ENST00000467793.5 |
RARRES2
|
retinoic acid receptor responder 2 |
| chr5_+_136058849 | 0.32 |
ENST00000508076.5
|
TGFBI
|
transforming growth factor beta induced |
| chr1_+_44746401 | 0.31 |
ENST00000372217.5
|
KIF2C
|
kinesin family member 2C |
| chr3_+_38165484 | 0.31 |
ENST00000446845.5
ENST00000311806.8 |
OXSR1
|
oxidative stress responsive kinase 1 |
| chr19_-_9858139 | 0.31 |
ENST00000590841.5
|
OLFM2
|
olfactomedin 2 |
| chr8_-_18808837 | 0.31 |
ENST00000286485.12
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr5_-_150086511 | 0.30 |
ENST00000675795.1
|
CSF1R
|
colony stimulating factor 1 receptor |
| chr9_+_27109393 | 0.30 |
ENST00000406359.8
|
TEK
|
TEK receptor tyrosine kinase |
| chr6_-_28443463 | 0.30 |
ENST00000289788.4
|
ZSCAN23
|
zinc finger and SCAN domain containing 23 |
| chr11_-_45918014 | 0.30 |
ENST00000525192.5
|
PEX16
|
peroxisomal biogenesis factor 16 |
| chr11_+_57598184 | 0.30 |
ENST00000677625.1
ENST00000676670.1 |
SERPING1
|
serpin family G member 1 |
| chr5_+_150166790 | 0.30 |
ENST00000616154.1
|
CDX1
|
caudal type homeobox 1 |
| chr19_+_35138778 | 0.29 |
ENST00000351325.9
ENST00000586871.5 ENST00000592174.1 |
FXYD1
ENSG00000221857.7
|
FXYD domain containing ion transport regulator 1 novel transcript |
| chr11_+_7513966 | 0.29 |
ENST00000299492.9
|
PPFIBP2
|
PPFIA binding protein 2 |
| chr1_+_153628393 | 0.29 |
ENST00000368696.3
ENST00000292169.6 ENST00000436839.1 |
S100A1
|
S100 calcium binding protein A1 |
| chr17_-_28951443 | 0.29 |
ENST00000268756.7
ENST00000332830.9 ENST00000584685.1 |
PHF12
|
PHD finger protein 12 |
| chr19_-_55370455 | 0.29 |
ENST00000264563.7
ENST00000585513.1 ENST00000590625.5 |
IL11
|
interleukin 11 |
| chr17_+_42798857 | 0.29 |
ENST00000588408.6
|
CNTD1
|
cyclin N-terminal domain containing 1 |
| chr5_-_59430600 | 0.29 |
ENST00000636120.1
|
PDE4D
|
phosphodiesterase 4D |
| chr1_+_44739825 | 0.29 |
ENST00000372224.9
|
KIF2C
|
kinesin family member 2C |
| chr17_+_42798881 | 0.29 |
ENST00000588527.5
|
CNTD1
|
cyclin N-terminal domain containing 1 |
| chr7_+_148590760 | 0.29 |
ENST00000307003.3
|
C7orf33
|
chromosome 7 open reading frame 33 |
| chr9_+_137230757 | 0.29 |
ENST00000673865.1
ENST00000538474.5 ENST00000673835.1 ENST00000673953.1 ENST00000361134.2 |
SLC34A3
|
solute carrier family 34 member 3 |
| chr5_+_147878703 | 0.29 |
ENST00000296694.5
|
SCGB3A2
|
secretoglobin family 3A member 2 |
| chr19_+_12838437 | 0.28 |
ENST00000251472.9
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr9_-_69759994 | 0.28 |
ENST00000340434.5
|
PTAR1
|
protein prenyltransferase alpha subunit repeat containing 1 |
| chr10_+_68109433 | 0.28 |
ENST00000613327.4
ENST00000358913.10 ENST00000373675.3 |
MYPN
|
myopalladin |
| chr10_+_69630227 | 0.28 |
ENST00000373279.6
|
FAM241B
|
family with sequence similarity 241 member B |
| chr14_+_93927259 | 0.28 |
ENST00000556222.1
ENST00000554404.1 |
FAM181A
|
family with sequence similarity 181 member A |
| chr12_-_132956280 | 0.28 |
ENST00000536932.5
ENST00000360187.9 ENST00000392321.3 |
CHFR
ZNF605
|
checkpoint with forkhead and ring finger domains zinc finger protein 605 |
| chr3_-_168095344 | 0.28 |
ENST00000309027.4
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr9_-_34665985 | 0.28 |
ENST00000416454.5
ENST00000544078.2 ENST00000421828.7 ENST00000423809.5 |
ENSG00000187186.15
|
novel protein |
| chr13_-_80341100 | 0.28 |
ENST00000377104.4
|
SPRY2
|
sprouty RTK signaling antagonist 2 |
| chr15_-_89211803 | 0.28 |
ENST00000563254.1
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr12_-_110920568 | 0.28 |
ENST00000548438.1
ENST00000228841.15 |
MYL2
|
myosin light chain 2 |
| chr10_-_118046574 | 0.27 |
ENST00000369199.5
|
RAB11FIP2
|
RAB11 family interacting protein 2 |
| chr12_+_51424802 | 0.27 |
ENST00000453097.7
|
SLC4A8
|
solute carrier family 4 member 8 |
| chr7_-_56092932 | 0.27 |
ENST00000446428.5
ENST00000432123.5 ENST00000297373.7 |
PHKG1
|
phosphorylase kinase catalytic subunit gamma 1 |
| chr8_-_92103217 | 0.27 |
ENST00000615601.4
ENST00000523629.5 |
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr3_-_24494875 | 0.27 |
ENST00000644321.1
|
THRB
|
thyroid hormone receptor beta |
| chr9_-_41128681 | 0.27 |
ENST00000622588.2
|
FOXD4L6
|
forkhead box D4 like 6 |
| chr22_+_31122923 | 0.27 |
ENST00000620191.4
ENST00000412277.6 ENST00000412985.5 ENST00000331075.10 ENST00000420017.5 ENST00000400294.6 ENST00000405300.5 ENST00000404390.7 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr3_+_50269140 | 0.27 |
ENST00000616701.5
ENST00000433753.4 ENST00000611067.4 |
SEMA3B
|
semaphorin 3B |
| chr19_+_50790415 | 0.27 |
ENST00000270593.2
|
ACP4
|
acid phosphatase 4 |
| chr19_+_45499797 | 0.26 |
ENST00000401593.5
ENST00000396736.2 |
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent 1N (putative) |
| chr19_-_48364034 | 0.26 |
ENST00000435956.7
|
TMEM143
|
transmembrane protein 143 |
| chr1_+_163069353 | 0.26 |
ENST00000531057.5
ENST00000527809.5 ENST00000367908.8 ENST00000367909.11 |
RGS4
|
regulator of G protein signaling 4 |
| chr1_-_203175783 | 0.26 |
ENST00000621380.1
ENST00000255416.9 |
MYBPH
|
myosin binding protein H |
| chrX_-_111410420 | 0.26 |
ENST00000371993.7
ENST00000680476.1 |
DCX
|
doublecortin |
| chr3_-_48088800 | 0.26 |
ENST00000423088.5
|
MAP4
|
microtubule associated protein 4 |
| chr22_+_20774092 | 0.26 |
ENST00000215727.10
|
SERPIND1
|
serpin family D member 1 |
| chr11_-_27700447 | 0.26 |
ENST00000356660.9
|
BDNF
|
brain derived neurotrophic factor |
| chr17_-_2025289 | 0.26 |
ENST00000331238.7
|
RTN4RL1
|
reticulon 4 receptor like 1 |
| chrX_+_71144818 | 0.26 |
ENST00000536169.5
ENST00000358741.4 ENST00000395855.6 ENST00000374051.7 |
NLGN3
|
neuroligin 3 |
| chr10_+_52314272 | 0.25 |
ENST00000373970.4
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr2_-_135837170 | 0.25 |
ENST00000264162.7
|
LCT
|
lactase |
| chr16_+_46884675 | 0.25 |
ENST00000562132.5
ENST00000440783.2 |
GPT2
|
glutamic--pyruvic transaminase 2 |
| chr1_-_46551647 | 0.25 |
ENST00000481882.7
|
KNCN
|
kinocilin |
| chr9_-_34381531 | 0.25 |
ENST00000379124.5
ENST00000379126.7 ENST00000379127.1 |
C9orf24
|
chromosome 9 open reading frame 24 |
| chr10_-_118046922 | 0.24 |
ENST00000355624.8
|
RAB11FIP2
|
RAB11 family interacting protein 2 |
| chr7_+_23710203 | 0.24 |
ENST00000422637.5
ENST00000355870.8 |
STK31
|
serine/threonine kinase 31 |
| chr7_+_23710326 | 0.24 |
ENST00000354639.7
ENST00000531170.5 ENST00000444333.2 |
STK31
|
serine/threonine kinase 31 |
| chr19_-_50163175 | 0.24 |
ENST00000293405.7
|
IZUMO2
|
IZUMO family member 2 |
| chr1_+_160177386 | 0.24 |
ENST00000470705.1
|
ATP1A4
|
ATPase Na+/K+ transporting subunit alpha 4 |
| chr15_+_45129933 | 0.24 |
ENST00000321429.8
ENST00000389037.7 ENST00000558322.5 |
DUOX1
|
dual oxidase 1 |
| chr9_+_72577788 | 0.24 |
ENST00000645208.2
|
TMC1
|
transmembrane channel like 1 |
| chr5_-_88268801 | 0.24 |
ENST00000506536.5
ENST00000512429.5 ENST00000514135.5 ENST00000296595.11 ENST00000509387.5 |
TMEM161B
|
transmembrane protein 161B |
| chr19_-_4517600 | 0.24 |
ENST00000301286.4
|
PLIN4
|
perilipin 4 |
| chr6_-_168093012 | 0.23 |
ENST00000646385.1
|
FRMD1
|
FERM domain containing 1 |
| chr17_+_7580442 | 0.23 |
ENST00000584180.1
|
CD68
|
CD68 molecule |
| chr14_+_93927358 | 0.23 |
ENST00000557000.2
|
FAM181A
|
family with sequence similarity 181 member A |
| chr17_-_74546082 | 0.23 |
ENST00000330793.2
|
CD300C
|
CD300c molecule |
| chr8_+_123072667 | 0.23 |
ENST00000519418.5
ENST00000287380.6 ENST00000327098.9 ENST00000522420.5 ENST00000521676.5 |
TBC1D31
|
TBC1 domain family member 31 |
| chr8_+_133191060 | 0.23 |
ENST00000519433.1
ENST00000517423.5 ENST00000220856.6 |
CCN4
|
cellular communication network factor 4 |
| chr17_-_28951285 | 0.23 |
ENST00000577226.5
|
PHF12
|
PHD finger protein 12 |
| chr19_+_14028148 | 0.23 |
ENST00000431365.3
ENST00000585987.1 |
RLN3
|
relaxin 3 |
| chr10_+_60778490 | 0.23 |
ENST00000448257.6
ENST00000614696.4 |
CDK1
|
cyclin dependent kinase 1 |
| chr11_-_40294089 | 0.23 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr2_-_27890348 | 0.23 |
ENST00000302188.8
|
RBKS
|
ribokinase |
| chr10_+_60778331 | 0.23 |
ENST00000519078.6
ENST00000316629.8 ENST00000395284.8 |
CDK1
|
cyclin dependent kinase 1 |
| chr9_+_72616266 | 0.23 |
ENST00000340019.4
|
TMC1
|
transmembrane channel like 1 |
| chr19_-_8502621 | 0.23 |
ENST00000600262.1
ENST00000423345.5 |
PRAM1
|
PML-RARA regulated adaptor molecule 1 |
| chr1_-_23559490 | 0.23 |
ENST00000374561.6
|
ID3
|
inhibitor of DNA binding 3, HLH protein |
| chr5_-_77638713 | 0.23 |
ENST00000306422.5
|
OTP
|
orthopedia homeobox |
| chr19_+_55376818 | 0.23 |
ENST00000291934.4
|
TMEM190
|
transmembrane protein 190 |
| chr16_+_2830368 | 0.23 |
ENST00000572863.1
|
ZG16B
|
zymogen granule protein 16B |
| chr4_+_105146868 | 0.22 |
ENST00000380013.9
ENST00000413648.2 |
TET2
|
tet methylcytosine dioxygenase 2 |
| chr1_+_171257930 | 0.22 |
ENST00000354841.4
|
FMO1
|
flavin containing dimethylaniline monoxygenase 1 |
| chr11_-_59845496 | 0.22 |
ENST00000257248.3
|
CBLIF
|
cobalamin binding intrinsic factor |
| chr20_+_45857607 | 0.22 |
ENST00000255152.3
|
ZSWIM3
|
zinc finger SWIM-type containing 3 |
| chrX_+_47080855 | 0.22 |
ENST00000336169.3
|
RGN
|
regucalcin |
| chr1_+_26159071 | 0.22 |
ENST00000374268.5
|
FAM110D
|
family with sequence similarity 110 member D |
| chr2_-_163735989 | 0.22 |
ENST00000333129.4
ENST00000409634.5 |
FIGN
|
fidgetin, microtubule severing factor |
| chr17_-_74868616 | 0.22 |
ENST00000579893.1
ENST00000544854.5 |
FDXR
|
ferredoxin reductase |
| chrX_+_13569593 | 0.22 |
ENST00000361306.6
ENST00000380602.3 |
EGFL6
|
EGF like domain multiple 6 |
| chr3_-_179072205 | 0.22 |
ENST00000432729.5
|
ZMAT3
|
zinc finger matrin-type 3 |
| chr6_-_30161200 | 0.22 |
ENST00000449742.7
|
TRIM10
|
tripartite motif containing 10 |
| chr22_+_50738198 | 0.22 |
ENST00000216139.10
ENST00000529621.1 |
ACR
|
acrosin |
| chr12_-_54300974 | 0.22 |
ENST00000435572.7
ENST00000553070.5 |
NFE2
|
nuclear factor, erythroid 2 |
| chr3_-_194351290 | 0.22 |
ENST00000429275.1
ENST00000323830.4 |
CPN2
|
carboxypeptidase N subunit 2 |
| chr11_-_116792386 | 0.21 |
ENST00000433069.2
ENST00000542499.5 |
APOA5
|
apolipoprotein A5 |
| chr9_-_21368057 | 0.21 |
ENST00000449498.2
|
IFNA13
|
interferon alpha 13 |
| chr4_+_663696 | 0.21 |
ENST00000471824.6
|
PDE6B
|
phosphodiesterase 6B |
| chr17_-_2711633 | 0.21 |
ENST00000435359.5
|
CLUH
|
clustered mitochondria homolog |
| chrX_+_16650155 | 0.21 |
ENST00000380200.3
|
S100G
|
S100 calcium binding protein G |
| chr16_+_56935371 | 0.21 |
ENST00000568358.1
|
HERPUD1
|
homocysteine inducible ER protein with ubiquitin like domain 1 |
| chr11_+_57597563 | 0.21 |
ENST00000619430.2
ENST00000457869.1 ENST00000340687.10 ENST00000278407.9 ENST00000378323.8 ENST00000378324.6 ENST00000403558.1 |
SERPING1
|
serpin family G member 1 |
| chrX_-_152451273 | 0.21 |
ENST00000370314.9
|
GABRA3
|
gamma-aminobutyric acid type A receptor subunit alpha3 |
| chr1_+_6555301 | 0.21 |
ENST00000333172.11
ENST00000351136.7 |
TAS1R1
|
taste 1 receptor member 1 |
| chr4_-_40515967 | 0.21 |
ENST00000381795.10
|
RBM47
|
RNA binding motif protein 47 |
| chr21_-_14658812 | 0.21 |
ENST00000647101.1
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr4_+_89111521 | 0.21 |
ENST00000603357.3
|
TIGD2
|
tigger transposable element derived 2 |
| chr16_+_30699155 | 0.21 |
ENST00000262518.9
|
SRCAP
|
Snf2 related CREBBP activator protein |
| chr16_-_31428325 | 0.20 |
ENST00000287490.5
|
COX6A2
|
cytochrome c oxidase subunit 6A2 |
| chr14_-_21536884 | 0.20 |
ENST00000546363.5
|
SALL2
|
spalt like transcription factor 2 |
| chr11_+_64340191 | 0.20 |
ENST00000356786.10
|
CCDC88B
|
coiled-coil domain containing 88B |
| chr21_+_44353607 | 0.20 |
ENST00000397928.6
|
TRPM2
|
transient receptor potential cation channel subfamily M member 2 |
| chr19_+_15640880 | 0.20 |
ENST00000586182.6
ENST00000221307.13 ENST00000591058.5 |
CYP4F3
|
cytochrome P450 family 4 subfamily F member 3 |
| chr1_-_27366917 | 0.20 |
ENST00000357582.3
|
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr14_-_104953899 | 0.20 |
ENST00000557457.1
|
AHNAK2
|
AHNAK nucleoprotein 2 |
| chr2_+_230996115 | 0.20 |
ENST00000424440.5
ENST00000452881.5 ENST00000433428.6 ENST00000455816.1 ENST00000440792.5 ENST00000423134.1 |
SPATA3
|
spermatogenesis associated 3 |
| chr12_-_54295748 | 0.20 |
ENST00000540264.2
ENST00000312156.8 |
NFE2
|
nuclear factor, erythroid 2 |
| chr12_+_108880085 | 0.20 |
ENST00000228476.8
ENST00000547768.5 |
DAO
|
D-amino acid oxidase |
| chr5_-_1886938 | 0.20 |
ENST00000613726.4
|
IRX4
|
iroquois homeobox 4 |
| chr19_-_49061997 | 0.20 |
ENST00000593537.1
|
NTF4
|
neurotrophin 4 |
| chrX_-_54798253 | 0.20 |
ENST00000218436.7
|
ITIH6
|
inter-alpha-trypsin inhibitor heavy chain family member 6 |
| chr9_+_137217452 | 0.19 |
ENST00000645271.1
|
ENSG00000284976.1
|
novel RING finger protein |
| chr21_+_44353633 | 0.19 |
ENST00000397932.6
ENST00000300481.13 |
TRPM2
|
transient receptor potential cation channel subfamily M member 2 |
| chr17_+_7558296 | 0.19 |
ENST00000438470.5
ENST00000436057.5 |
TNFSF13
|
TNF superfamily member 13 |
| chr14_+_88385643 | 0.19 |
ENST00000393545.9
ENST00000356583.9 ENST00000555401.5 ENST00000553885.5 |
SPATA7
|
spermatogenesis associated 7 |
| chr6_-_41154326 | 0.19 |
ENST00000426005.6
ENST00000437044.2 ENST00000373127.8 |
TREML1
|
triggering receptor expressed on myeloid cells like 1 |
| chr21_+_42403856 | 0.19 |
ENST00000291535.11
|
UBASH3A
|
ubiquitin associated and SH3 domain containing A |
| chr7_-_56092974 | 0.19 |
ENST00000452681.6
ENST00000537360.5 |
PHKG1
|
phosphorylase kinase catalytic subunit gamma 1 |
| chr8_-_26513865 | 0.19 |
ENST00000522362.7
|
PNMA2
|
PNMA family member 2 |
| chr19_+_35030438 | 0.19 |
ENST00000415950.5
ENST00000262631.11 |
SCN1B
|
sodium voltage-gated channel beta subunit 1 |
| chr7_-_73624492 | 0.19 |
ENST00000414749.6
ENST00000429400.6 ENST00000434326.5 ENST00000313375.8 ENST00000354613.5 ENST00000453275.1 |
MLXIPL
|
MLX interacting protein like |
| chr19_+_6464229 | 0.19 |
ENST00000600229.6
ENST00000356762.7 |
CRB3
|
crumbs cell polarity complex component 3 |
| chr8_+_37695774 | 0.19 |
ENST00000331569.6
|
ZNF703
|
zinc finger protein 703 |
| chrX_+_47078380 | 0.19 |
ENST00000352078.8
|
RGN
|
regucalcin |
| chrX_+_47078330 | 0.19 |
ENST00000457380.5
|
RGN
|
regucalcin |
| chr6_-_30160880 | 0.19 |
ENST00000376704.3
|
TRIM10
|
tripartite motif containing 10 |
| chr14_-_72894091 | 0.19 |
ENST00000556509.6
|
DPF3
|
double PHD fingers 3 |
| chr18_-_49813512 | 0.18 |
ENST00000285093.15
|
ACAA2
|
acetyl-CoA acyltransferase 2 |
| chr9_+_68302867 | 0.18 |
ENST00000342833.4
|
FOXD4L3
|
forkhead box D4 like 3 |
| chr12_-_91179355 | 0.18 |
ENST00000550563.5
ENST00000546370.5 |
DCN
|
decorin |
| chr17_-_5486157 | 0.18 |
ENST00000572834.5
ENST00000158771.9 ENST00000570848.5 ENST00000571971.1 |
DERL2
|
derlin 2 |
| chr19_-_47419490 | 0.18 |
ENST00000331559.9
ENST00000558555.6 |
MEIS3
|
Meis homeobox 3 |
| chr9_+_117704382 | 0.18 |
ENST00000646089.1
ENST00000355622.8 |
ENSG00000285082.2
TLR4
|
novel protein toll like receptor 4 |
| chr18_+_61333424 | 0.18 |
ENST00000262717.9
|
CDH20
|
cadherin 20 |
| chr11_-_58575846 | 0.18 |
ENST00000395074.7
|
LPXN
|
leupaxin |
| chr10_+_68560317 | 0.18 |
ENST00000373644.5
|
TET1
|
tet methylcytosine dioxygenase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0007225 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.2 | 0.6 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.2 | 0.5 | GO:0085032 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.2 | 0.5 | GO:1900920 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.1 | 0.6 | GO:1903631 | regulation of calcium-dependent ATPase activity(GO:1903610) negative regulation of calcium-dependent ATPase activity(GO:1903611) regulation of dUTP diphosphatase activity(GO:1903627) positive regulation of dUTP diphosphatase activity(GO:1903629) negative regulation of aminoacyl-tRNA ligase activity(GO:1903631) regulation of leucine-tRNA ligase activity(GO:1903633) negative regulation of leucine-tRNA ligase activity(GO:1903634) |
| 0.1 | 0.4 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.1 | 0.4 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.1 | 0.5 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.1 | 0.4 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.1 | 0.3 | GO:0030718 | inner cell mass cell fate commitment(GO:0001827) germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.3 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.1 | 0.7 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 0.5 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.8 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 0.5 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.1 | 0.3 | GO:2000374 | regulation of oxygen metabolic process(GO:2000374) |
| 0.1 | 0.3 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.1 | 0.3 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 0.2 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.6 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.4 | GO:0006524 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.1 | 0.3 | GO:0060437 | lung growth(GO:0060437) |
| 0.1 | 0.2 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.1 | 0.3 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 0.2 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.1 | 0.3 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.6 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.4 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.4 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.4 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.2 | GO:0048560 | establishment of anatomical structure orientation(GO:0048560) |
| 0.1 | 0.5 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 0.3 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.1 | 0.4 | GO:0036102 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
| 0.1 | 0.6 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 0.5 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.1 | 0.2 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.1 | 0.2 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 0.2 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.1 | 0.2 | GO:0061075 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.1 | 0.3 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.1 | 0.2 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.1 | 0.2 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.1 | 0.3 | GO:0019477 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.1 | 0.1 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.1 | 0.2 | GO:0003290 | atrial septum secundum morphogenesis(GO:0003290) embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.0 | GO:0019878 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.0 | 0.2 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.3 | GO:0010731 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.0 | 0.1 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.1 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.5 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0051695 | actin filament uncapping(GO:0051695) |
| 0.0 | 0.2 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.0 | 0.1 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.0 | 0.4 | GO:2000317 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.0 | 0.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.3 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.1 | GO:0009946 | inhibition of neuroepithelial cell differentiation(GO:0002085) proximal/distal axis specification(GO:0009946) |
| 0.0 | 0.4 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.0 | 0.4 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.2 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.1 | GO:0051939 | gamma-aminobutyric acid import(GO:0051939) |
| 0.0 | 0.1 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.5 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.5 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.1 | GO:1904170 | regulation of bleb assembly(GO:1904170) positive regulation of bleb assembly(GO:1904172) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.0 | 0.1 | GO:0060032 | notochord regression(GO:0060032) |
| 0.0 | 0.1 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 0.1 | GO:1902269 | putrescine transport(GO:0015847) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.4 | GO:2000620 | innate vocalization behavior(GO:0098582) positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.2 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.1 | GO:1903937 | response to acrylamide(GO:1903937) |
| 0.0 | 0.2 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.1 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.0 | 0.3 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.1 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.9 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.3 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.4 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:2001302 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.0 | 0.2 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.2 | GO:0002840 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) |
| 0.0 | 0.1 | GO:1904253 | circadian temperature homeostasis(GO:0060086) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.0 | 0.1 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.1 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.2 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.5 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.2 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.3 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 0.1 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.2 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0043449 | dibenzo-p-dioxin metabolic process(GO:0018894) cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.1 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.0 | 0.2 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) lobar bronchus development(GO:0060482) |
| 0.0 | 0.1 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.6 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.2 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.1 | GO:0019483 | beta-alanine biosynthetic process(GO:0019483) |
| 0.0 | 0.1 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.2 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.2 | GO:0060694 | regulation of cholesterol transporter activity(GO:0060694) pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.0 | GO:0072040 | regulation of mesenchymal cell apoptotic process involved in nephron morphogenesis(GO:0072039) negative regulation of mesenchymal cell apoptotic process involved in nephron morphogenesis(GO:0072040) mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) mesenchymal cell apoptotic process involved in nephron morphogenesis(GO:1901145) negative regulation of somatic stem cell population maintenance(GO:1904673) |
| 0.0 | 0.1 | GO:0052031 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.0 | 0.1 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.0 | 0.0 | GO:0007418 | ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.0 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.0 | 0.2 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.1 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0046081 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.0 | 0.1 | GO:1902769 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.3 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.2 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.1 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.0 | 0.2 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) vagus nerve development(GO:0021564) |
| 0.0 | 0.0 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) |
| 0.0 | 0.1 | GO:0090027 | negative regulation of monocyte chemotaxis(GO:0090027) |
| 0.0 | 0.2 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.2 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.2 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.0 | 0.3 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.1 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.0 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.1 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.0 | 0.1 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 | 0.4 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.4 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.1 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.2 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.0 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.1 | GO:0019364 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.0 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.0 | 0.1 | GO:0033967 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.2 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.1 | GO:2000724 | mineralocorticoid receptor signaling pathway(GO:0031959) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0090299 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.0 | 0.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.1 | GO:1903182 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.2 | GO:0003072 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.0 | 0.4 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.1 | GO:0043504 | DNA replication, removal of RNA primer(GO:0043137) mitochondrial DNA repair(GO:0043504) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 0.2 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.0 | 0.1 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.0 | 0.3 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.2 | GO:0010255 | hexose mediated signaling(GO:0009757) sugar mediated signaling pathway(GO:0010182) glucose mediated signaling pathway(GO:0010255) |
| 0.0 | 0.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.1 | GO:0072268 | nephrogenic mesenchyme development(GO:0072076) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.2 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.3 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.0 | 0.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.5 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.1 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.1 | GO:0071626 | mastication(GO:0071626) learned vocalization behavior(GO:0098583) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.2 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.1 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.3 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.0 | 0.1 | GO:2000612 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.1 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.4 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.0 | 0.1 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0046449 | creatinine metabolic process(GO:0046449) |
| 0.0 | 0.2 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.1 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.0 | 0.2 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.2 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.2 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.5 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.2 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.1 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.3 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.1 | GO:0021936 | regulation of cerebellar granule cell precursor proliferation(GO:0021936) |
| 0.0 | 0.1 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.0 | GO:0052572 | response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.0 | 0.2 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.0 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 2.1 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.0 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.0 | 0.1 | GO:2001270 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) |
| 0.0 | 0.1 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.0 | 0.1 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.0 | GO:0014810 | positive regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014810) |
| 0.0 | 0.0 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.0 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.2 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.4 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:1903352 | L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.1 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.0 | 0.0 | GO:0002584 | negative regulation of antigen processing and presentation of peptide antigen(GO:0002584) |
| 0.0 | 0.1 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.0 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.3 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.3 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.2 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.1 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 0.1 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.0 | 0.7 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.2 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.1 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.0 | GO:0032847 | regulation of cellular pH reduction(GO:0032847) negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.3 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.0 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.2 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.0 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.0 | 0.2 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.1 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.0 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.1 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.0 | 0.5 | GO:0003091 | renal water homeostasis(GO:0003091) |
| 0.0 | 0.0 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:0002638 | negative regulation of immunoglobulin production(GO:0002638) |
| 0.0 | 0.1 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.0 | 0.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.1 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.0 | 0.0 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.7 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.2 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 0.5 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.6 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.2 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.0 | 0.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.5 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.1 | GO:1990356 | sumoylated E2 ligase complex(GO:1990356) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.1 | GO:0097545 | axonemal outer doublet(GO:0097545) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.8 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 1.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.0 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.0 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.0 | 0.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.1 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.0 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.1 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0004341 | gluconolactonase activity(GO:0004341) |
| 0.1 | 1.0 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.4 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.3 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.1 | 0.6 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.3 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.1 | 0.3 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.1 | 0.2 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.1 | 0.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.6 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 0.3 | GO:0004040 | amidase activity(GO:0004040) |
| 0.1 | 1.5 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.1 | 0.5 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.2 | GO:0097258 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.1 | 0.3 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 0.2 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 0.2 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.1 | 0.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 0.3 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.1 | 0.3 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.7 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.3 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.2 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.0 | 0.5 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.0 | 0.2 | GO:0015056 | corticotrophin-releasing factor receptor activity(GO:0015056) |
| 0.0 | 0.3 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.2 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.0 | 0.1 | GO:0015203 | polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) |
| 0.0 | 0.2 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.2 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.1 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.2 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.5 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.7 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.6 | GO:0019841 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.5 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.3 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.6 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.0 | 0.5 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.0 | 0.1 | GO:0016890 | site-specific endodeoxyribonuclease activity, specific for altered base(GO:0016890) |
| 0.0 | 0.2 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.1 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.3 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0004750 | ribulose-phosphate 3-epimerase activity(GO:0004750) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.1 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0047750 | cholestenol delta-isomerase activity(GO:0047750) |
| 0.0 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.1 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.0 | 0.1 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.0 | 0.3 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0001034 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0016807 | cysteine-type carboxypeptidase activity(GO:0016807) cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.2 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.0 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.1 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.4 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.4 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.5 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.0 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.0 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.3 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.0 | 0.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.0 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.0 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.0 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.0 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.0 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 1.2 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.0 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.5 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.6 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 1.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.8 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |