|
chr6_-_29559724
Show fit
|
10.99 |
ENST00000377050.5
|
UBD
|
ubiquitin D
|
|
chr9_-_120926752
Show fit
|
7.64 |
ENST00000373887.8
|
TRAF1
|
TNF receptor associated factor 1
|
|
chr1_-_173050931
Show fit
|
7.06 |
ENST00000404377.5
|
TNFSF18
|
TNF superfamily member 18
|
|
chr4_+_73836667
Show fit
|
6.72 |
ENST00000226317.10
|
CXCL6
|
C-X-C motif chemokine ligand 6
|
|
chr1_-_7940825
Show fit
|
6.51 |
ENST00000377507.8
|
TNFRSF9
|
TNF receptor superfamily member 9
|
|
chr11_+_102317492
Show fit
|
5.08 |
ENST00000673846.1
|
BIRC3
|
baculoviral IAP repeat containing 3
|
|
chr1_+_109910485
Show fit
|
4.97 |
ENST00000525659.5
|
CSF1
|
colony stimulating factor 1
|
|
chr14_+_103123452
Show fit
|
4.91 |
ENST00000558056.1
ENST00000560869.6
|
TNFAIP2
|
TNF alpha induced protein 2
|
|
chr6_+_14117764
Show fit
|
4.82 |
ENST00000379153.4
|
CD83
|
CD83 molecule
|
|
chr12_+_77830886
Show fit
|
4.66 |
ENST00000397909.7
ENST00000549464.5
|
NAV3
|
neuron navigator 3
|
|
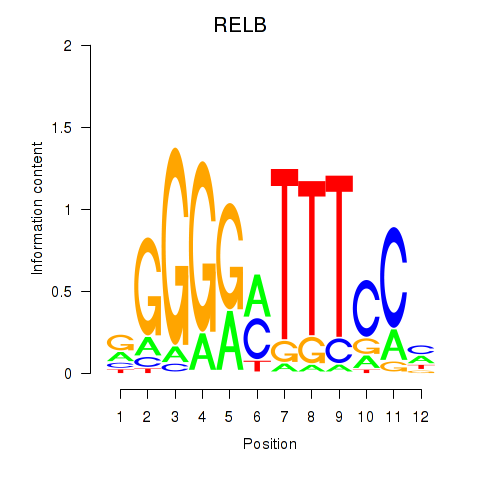
chr19_+_45001430
Show fit
|
4.62 |
ENST00000625761.2
ENST00000505236.1
ENST00000221452.13
|
RELB
|
RELB proto-oncogene, NF-kB subunit
|
|
chr4_-_73998669
Show fit
|
4.54 |
ENST00000296027.5
|
CXCL5
|
C-X-C motif chemokine ligand 5
|
|
chr4_+_102501885
Show fit
|
4.28 |
ENST00000505458.5
|
NFKB1
|
nuclear factor kappa B subunit 1
|
|
chr5_-_151080978
Show fit
|
4.00 |
ENST00000520931.5
ENST00000521591.6
ENST00000520695.5
ENST00000610535.5
ENST00000518977.5
ENST00000389378.6
ENST00000610874.4
|
TNIP1
|
TNFAIP3 interacting protein 1
|
|
chr5_+_10564064
Show fit
|
3.91 |
ENST00000296657.7
|
ANKRD33B
|
ankyrin repeat domain 33B
|
|
chr1_+_109910840
Show fit
|
3.88 |
ENST00000329608.11
ENST00000488198.5
|
CSF1
|
colony stimulating factor 1
|
|
chr6_+_32854179
Show fit
|
3.75 |
ENST00000374859.3
|
PSMB9
|
proteasome 20S subunit beta 9
|
|
chr11_+_102317542
Show fit
|
3.70 |
ENST00000532808.5
|
BIRC3
|
baculoviral IAP repeat containing 3
|
|
chr11_+_102317450
Show fit
|
3.64 |
ENST00000615299.4
ENST00000527309.2
ENST00000526421.6
ENST00000263464.9
|
BIRC3
|
baculoviral IAP repeat containing 3
|
|
chr10_+_102394488
Show fit
|
3.54 |
ENST00000369966.8
|
NFKB2
|
nuclear factor kappa B subunit 2
|
|
chr12_-_48865863
Show fit
|
3.53 |
ENST00000309739.6
|
RND1
|
Rho family GTPase 1
|
|
chr6_-_142946312
Show fit
|
3.36 |
ENST00000367604.6
|
HIVEP2
|
HIVEP zinc finger 2
|
|
chr2_-_162318475
Show fit
|
3.26 |
ENST00000648433.1
|
IFIH1
|
interferon induced with helicase C domain 1
|
|
chr6_+_137867241
Show fit
|
3.23 |
ENST00000612899.5
ENST00000420009.5
|
TNFAIP3
|
TNF alpha induced protein 3
|
|
chr6_+_137867414
Show fit
|
3.22 |
ENST00000237289.8
ENST00000433680.1
|
TNFAIP3
|
TNF alpha induced protein 3
|
|
chr9_-_133479075
Show fit
|
3.18 |
ENST00000414172.1
ENST00000371897.8
ENST00000371899.9
|
SLC2A6
|
solute carrier family 2 member 6
|
|
chr12_-_57110284
Show fit
|
2.97 |
ENST00000543873.6
ENST00000554663.5
ENST00000557635.5
|
STAT6
|
signal transducer and activator of transcription 6
|
|
chr5_-_147453888
Show fit
|
2.82 |
ENST00000398514.7
|
DPYSL3
|
dihydropyrimidinase like 3
|
|
chr17_+_42288464
Show fit
|
2.45 |
ENST00000590726.7
ENST00000678903.1
ENST00000590949.6
ENST00000676585.1
ENST00000588868.5
|
STAT5A
|
signal transducer and activator of transcription 5A
|
|
chr22_-_19525369
Show fit
|
2.28 |
ENST00000403084.1
ENST00000413119.2
|
CLDN5
|
claudin 5
|
|
chr17_+_42288429
Show fit
|
2.21 |
ENST00000676631.1
ENST00000677893.1
|
STAT5A
|
signal transducer and activator of transcription 5A
|
|
chr18_+_58149314
Show fit
|
2.20 |
ENST00000435432.6
ENST00000357895.9
ENST00000586263.5
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase
|
|
chr16_+_50696999
Show fit
|
2.11 |
ENST00000300589.6
|
NOD2
|
nucleotide binding oligomerization domain containing 2
|
|
chr4_-_184474518
Show fit
|
2.10 |
ENST00000393593.8
|
IRF2
|
interferon regulatory factor 2
|
|
chr8_+_53880867
Show fit
|
2.09 |
ENST00000522225.5
|
RGS20
|
regulator of G protein signaling 20
|
|
chr6_+_29723340
Show fit
|
2.08 |
ENST00000334668.8
|
HLA-F
|
major histocompatibility complex, class I, F
|
|
chr8_+_53880894
Show fit
|
2.02 |
ENST00000276500.4
|
RGS20
|
regulator of G protein signaling 20
|
|
chr6_+_29723421
Show fit
|
1.91 |
ENST00000259951.12
ENST00000434407.6
|
HLA-F
|
major histocompatibility complex, class I, F
|
|
chr21_+_33403466
Show fit
|
1.85 |
ENST00000405436.5
|
IFNGR2
|
interferon gamma receptor 2
|
|
chr19_-_4338786
Show fit
|
1.80 |
ENST00000601482.1
ENST00000600324.5
ENST00000594605.6
|
STAP2
|
signal transducing adaptor family member 2
|
|
chr2_-_203535253
Show fit
|
1.80 |
ENST00000457812.5
ENST00000319170.10
ENST00000630330.2
ENST00000308091.8
ENST00000453034.5
ENST00000420371.2
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1
|
|
chr2_+_68467544
Show fit
|
1.78 |
ENST00000303795.9
|
APLF
|
aprataxin and PNKP like factor
|
|
chr2_-_162318613
Show fit
|
1.77 |
ENST00000649979.2
ENST00000421365.2
|
IFIH1
|
interferon induced with helicase C domain 1
|
|
chr10_+_13100075
Show fit
|
1.76 |
ENST00000378747.8
ENST00000378757.6
ENST00000378752.7
ENST00000378748.7
|
OPTN
|
optineurin
|
|
chr10_+_13099585
Show fit
|
1.72 |
ENST00000378764.6
|
OPTN
|
optineurin
|
|
chr2_+_32628026
Show fit
|
1.71 |
ENST00000448773.5
ENST00000317907.9
|
TTC27
|
tetratricopeptide repeat domain 27
|
|
chr10_+_12349533
Show fit
|
1.70 |
ENST00000619168.5
|
CAMK1D
|
calcium/calmodulin dependent protein kinase ID
|
|
chr2_+_26346086
Show fit
|
1.68 |
ENST00000613142.4
ENST00000260585.12
ENST00000447170.1
|
SELENOI
|
selenoprotein I
|
|
chr4_+_73869385
Show fit
|
1.57 |
ENST00000395761.4
|
CXCL1
|
C-X-C motif chemokine ligand 1
|
|
chr9_-_32526185
Show fit
|
1.47 |
ENST00000379883.3
ENST00000379868.6
ENST00000679859.1
|
DDX58
|
DExD/H-box helicase 58
|
|
chr6_+_29942523
Show fit
|
1.45 |
ENST00000376809.10
ENST00000376802.2
ENST00000638375.1
|
HLA-A
|
major histocompatibility complex, class I, A
|
|
chr1_+_159204860
Show fit
|
1.41 |
ENST00000368122.4
ENST00000368121.6
|
ACKR1
|
atypical chemokine receptor 1 (Duffy blood group)
|
|
chr10_+_94545852
Show fit
|
1.39 |
ENST00000394036.5
ENST00000394045.5
|
HELLS
|
helicase, lymphoid specific
|
|
chr5_-_59893718
Show fit
|
1.39 |
ENST00000340635.11
|
PDE4D
|
phosphodiesterase 4D
|
|
chr2_-_96505345
Show fit
|
1.38 |
ENST00000310865.7
ENST00000451794.6
|
NEURL3
|
neuralized E3 ubiquitin protein ligase 3
|
|
chr1_-_64966488
Show fit
|
1.36 |
ENST00000342505.5
ENST00000673254.1
|
JAK1
|
Janus kinase 1
|
|
chr21_+_33403391
Show fit
|
1.35 |
ENST00000290219.11
ENST00000381995.5
|
IFNGR2
|
interferon gamma receptor 2
|
|
chr3_-_39153512
Show fit
|
1.28 |
ENST00000273153.10
|
CSRNP1
|
cysteine and serine rich nuclear protein 1
|
|
chr8_+_144095054
Show fit
|
1.23 |
ENST00000318911.5
|
CYC1
|
cytochrome c1
|
|
chr10_-_73414027
Show fit
|
1.21 |
ENST00000372921.10
ENST00000372919.8
|
ANXA7
|
annexin A7
|
|
chr3_+_183253230
Show fit
|
1.21 |
ENST00000326505.4
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5
|
|
chr10_+_94545777
Show fit
|
1.19 |
ENST00000348459.10
ENST00000419900.5
ENST00000630929.2
|
HELLS
|
helicase, lymphoid specific
|
|
chr6_-_11779606
Show fit
|
1.11 |
ENST00000506810.1
|
ADTRP
|
androgen dependent TFPI regulating protein
|
|
chr2_-_100104530
Show fit
|
1.10 |
ENST00000432037.5
ENST00000673232.1
ENST00000423966.6
ENST00000409236.6
|
AFF3
|
AF4/FMR2 family member 3
|
|
chr12_-_54588516
Show fit
|
1.09 |
ENST00000547431.5
|
PPP1R1A
|
protein phosphatase 1 regulatory inhibitor subunit 1A
|
|
chr19_-_17847962
Show fit
|
1.06 |
ENST00000458235.7
ENST00000534444.1
|
JAK3
|
Janus kinase 3
|
|
chr7_+_116524984
Show fit
|
1.05 |
ENST00000614113.5
ENST00000393470.1
|
CAV1
|
caveolin 1
|
|
chr17_-_79009778
Show fit
|
1.03 |
ENST00000591773.5
ENST00000588611.5
ENST00000586916.6
ENST00000592033.5
ENST00000588075.5
ENST00000302345.6
ENST00000591811.1
|
CANT1
|
calcium activated nucleotidase 1
|
|
chr12_-_12562851
Show fit
|
1.03 |
ENST00000298573.9
|
DUSP16
|
dual specificity phosphatase 16
|
|
chrX_-_11665908
Show fit
|
1.01 |
ENST00000337414.9
|
ARHGAP6
|
Rho GTPase activating protein 6
|
|
chr7_+_116525524
Show fit
|
0.99 |
ENST00000405348.6
|
CAV1
|
caveolin 1
|
|
chr2_-_199955464
Show fit
|
0.95 |
ENST00000354611.9
|
TYW5
|
tRNA-yW synthesizing protein 5
|
|
chr11_-_5154757
Show fit
|
0.94 |
ENST00000380367.3
|
OR52A1
|
olfactory receptor family 52 subfamily A member 1
|
|
chr1_-_247078804
Show fit
|
0.93 |
ENST00000366503.3
|
ZNF670
|
zinc finger protein 670
|
|
chr6_-_136525961
Show fit
|
0.91 |
ENST00000438100.6
|
MAP7
|
microtubule associated protein 7
|
|
chr1_-_160343235
Show fit
|
0.91 |
ENST00000368069.7
ENST00000241704.8
ENST00000647683.1
ENST00000649787.1
|
COPA
|
COPI coat complex subunit alpha
|
|
chr2_+_207529731
Show fit
|
0.89 |
ENST00000430624.5
|
CREB1
|
cAMP responsive element binding protein 1
|
|
chrX_+_73447042
Show fit
|
0.89 |
ENST00000373514.3
|
CDX4
|
caudal type homeobox 4
|
|
chr6_-_36387566
Show fit
|
0.89 |
ENST00000339796.9
ENST00000627426.2
|
ETV7
|
ETS variant transcription factor 7
|
|
chr10_+_87659839
Show fit
|
0.88 |
ENST00000456849.2
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2
|
|
chr5_+_68288346
Show fit
|
0.87 |
ENST00000320694.12
|
PIK3R1
|
phosphoinositide-3-kinase regulatory subunit 1
|
|
chr17_-_7687427
Show fit
|
0.87 |
ENST00000514944.5
ENST00000503591.1
ENST00000610292.4
ENST00000420246.6
ENST00000455263.6
ENST00000610538.4
ENST00000622645.4
ENST00000445888.6
ENST00000619485.4
ENST00000509690.5
ENST00000604348.5
ENST00000269305.9
ENST00000620739.4
|
TP53
|
tumor protein p53
|
|
chr8_-_12755457
Show fit
|
0.86 |
ENST00000398246.8
|
LONRF1
|
LON peptidase N-terminal domain and ring finger 1
|
|
chr17_-_79009731
Show fit
|
0.86 |
ENST00000392446.10
ENST00000590370.5
ENST00000591625.5
|
CANT1
|
calcium activated nucleotidase 1
|
|
chr10_-_72215903
Show fit
|
0.86 |
ENST00000526751.5
ENST00000672774.1
ENST00000394919.5
ENST00000394915.7
|
ASCC1
|
activating signal cointegrator 1 complex subunit 1
|
|
chr15_-_101277287
Show fit
|
0.84 |
ENST00000528346.1
ENST00000531964.5
ENST00000398226.7
ENST00000526049.6
|
SELENOS
|
selenoprotein S
|
|
chr14_-_72894091
Show fit
|
0.83 |
ENST00000556509.6
|
DPF3
|
double PHD fingers 3
|
|
chr1_+_100538131
Show fit
|
0.82 |
ENST00000315033.5
|
GPR88
|
G protein-coupled receptor 88
|
|
chr10_-_72216267
Show fit
|
0.78 |
ENST00000342444.8
ENST00000533958.1
ENST00000672957.1
ENST00000527593.5
ENST00000672940.1
ENST00000530461.5
ENST00000317168.11
ENST00000524829.5
|
ASCC1
|
activating signal cointegrator 1 complex subunit 1
|
|
chr12_+_2053545
Show fit
|
0.78 |
ENST00000682336.1
ENST00000344100.7
ENST00000399591.5
ENST00000399595.5
ENST00000399597.5
ENST00000399601.5
ENST00000399606.5
ENST00000399621.5
ENST00000399629.5
ENST00000399637.5
ENST00000399638.5
ENST00000399644.5
ENST00000399649.5
ENST00000402845.7
ENST00000682686.1
|
CACNA1C
|
calcium voltage-gated channel subunit alpha1 C
|
|
chr1_-_153540694
Show fit
|
0.76 |
ENST00000368717.2
|
S100A5
|
S100 calcium binding protein A5
|
|
chr19_+_38899946
Show fit
|
0.76 |
ENST00000572515.5
ENST00000313582.6
ENST00000575359.5
|
NFKBIB
|
NFKB inhibitor beta
|
|
chr20_-_18497218
Show fit
|
0.75 |
ENST00000337227.9
|
RBBP9
|
RB binding protein 9, serine hydrolase
|
|
chr19_+_38899680
Show fit
|
0.75 |
ENST00000576510.5
ENST00000392079.7
|
NFKBIB
|
NFKB inhibitor beta
|
|
chr1_-_204411804
Show fit
|
0.75 |
ENST00000367188.5
|
PPP1R15B
|
protein phosphatase 1 regulatory subunit 15B
|
|
chr16_-_2009797
Show fit
|
0.75 |
ENST00000563630.6
ENST00000562103.2
|
ZNF598
|
zinc finger protein 598, E3 ubiquitin ligase
|
|
chr1_-_196608359
Show fit
|
0.74 |
ENST00000609185.5
ENST00000451324.6
ENST00000367433.9
ENST00000294725.14
|
KCNT2
|
potassium sodium-activated channel subfamily T member 2
|
|
chr2_-_27495185
Show fit
|
0.74 |
ENST00000264703.4
|
FNDC4
|
fibronectin type III domain containing 4
|
|
chr4_+_39044995
Show fit
|
0.74 |
ENST00000261425.7
ENST00000508137.6
|
KLHL5
|
kelch like family member 5
|
|
chr16_+_57358775
Show fit
|
0.73 |
ENST00000219235.5
|
CCL22
|
C-C motif chemokine ligand 22
|
|
chr10_+_94762673
Show fit
|
0.72 |
ENST00000480405.2
ENST00000371321.9
|
CYP2C19
|
cytochrome P450 family 2 subfamily C member 19
|
|
chr15_-_78944985
Show fit
|
0.70 |
ENST00000615999.5
ENST00000677789.1
ENST00000676880.1
ENST00000677936.1
ENST00000220166.10
ENST00000677810.1
ENST00000678644.1
ENST00000677534.1
ENST00000677316.1
|
CTSH
|
cathepsin H
|
|
chr1_+_25338294
Show fit
|
0.70 |
ENST00000374358.5
|
TMEM50A
|
transmembrane protein 50A
|
|
chr22_-_37244417
Show fit
|
0.69 |
ENST00000405484.5
ENST00000441619.5
ENST00000406508.5
|
RAC2
|
Rac family small GTPase 2
|
|
chr14_+_102777461
Show fit
|
0.68 |
ENST00000560371.5
ENST00000347662.8
|
TRAF3
|
TNF receptor associated factor 3
|
|
chr6_-_36387736
Show fit
|
0.68 |
ENST00000373737.8
ENST00000373738.4
ENST00000538992.3
ENST00000615781.4
ENST00000620358.4
|
ETV7
|
ETS variant transcription factor 7
|
|
chr4_+_4859658
Show fit
|
0.68 |
ENST00000382723.5
|
MSX1
|
msh homeobox 1
|
|
chr3_-_108222383
Show fit
|
0.68 |
ENST00000264538.4
|
IFT57
|
intraflagellar transport 57
|
|
chr7_-_22500152
Show fit
|
0.67 |
ENST00000406890.6
ENST00000678116.1
ENST00000424363.5
|
STEAP1B
|
STEAP family member 1B
|
|
chr14_+_102777555
Show fit
|
0.65 |
ENST00000539721.5
ENST00000560463.5
|
TRAF3
|
TNF receptor associated factor 3
|
|
chr11_+_72114840
Show fit
|
0.65 |
ENST00000622388.4
|
FOLR3
|
folate receptor gamma
|
|
chr6_-_136526472
Show fit
|
0.64 |
ENST00000454590.5
ENST00000432797.6
|
MAP7
|
microtubule associated protein 7
|
|
chr6_-_36387654
Show fit
|
0.64 |
ENST00000340181.9
|
ETV7
|
ETS variant transcription factor 7
|
|
chrX_+_15500800
Show fit
|
0.63 |
ENST00000348343.11
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr9_-_133348022
Show fit
|
0.63 |
ENST00000446777.5
ENST00000610888.4
ENST00000371999.5
ENST00000494177.6
ENST00000343730.10
ENST00000457204.2
|
MED22
|
mediator complex subunit 22
|
|
chr7_+_116525028
Show fit
|
0.62 |
ENST00000341049.7
|
CAV1
|
caveolin 1
|
|
chr12_-_124989053
Show fit
|
0.61 |
ENST00000308736.7
|
DHX37
|
DEAH-box helicase 37
|
|
chr4_+_77605807
Show fit
|
0.60 |
ENST00000682537.1
|
CXCL13
|
C-X-C motif chemokine ligand 13
|
|
chr2_+_64454506
Show fit
|
0.60 |
ENST00000409537.2
|
LGALSL
|
galectin like
|
|
chr16_-_57284654
Show fit
|
0.60 |
ENST00000613167.4
ENST00000219207.10
ENST00000569059.5
|
PLLP
|
plasmolipin
|
|
chr4_-_74099187
Show fit
|
0.60 |
ENST00000508487.3
|
CXCL2
|
C-X-C motif chemokine ligand 2
|
|
chr2_+_64453969
Show fit
|
0.59 |
ENST00000464281.5
|
LGALSL
|
galectin like
|
|
chr11_+_64284794
Show fit
|
0.59 |
ENST00000539851.5
|
GPR137
|
G protein-coupled receptor 137
|
|
chr6_-_35688907
Show fit
|
0.59 |
ENST00000539068.5
ENST00000357266.9
|
FKBP5
|
FKBP prolyl isomerase 5
|
|
chr1_+_109910892
Show fit
|
0.59 |
ENST00000369802.7
ENST00000420111.6
|
CSF1
|
colony stimulating factor 1
|
|
chr19_-_40218339
Show fit
|
0.59 |
ENST00000311308.6
|
TTC9B
|
tetratricopeptide repeat domain 9B
|
|
chr8_-_100950549
Show fit
|
0.58 |
ENST00000395951.7
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta
|
|
chr14_-_100568475
Show fit
|
0.58 |
ENST00000553553.6
|
BEGAIN
|
brain enriched guanylate kinase associated
|
|
chr15_-_30991415
Show fit
|
0.58 |
ENST00000563714.5
|
MTMR10
|
myotubularin related protein 10
|
|
chr10_+_84328625
Show fit
|
0.56 |
ENST00000224756.12
|
CCSER2
|
coiled-coil serine rich protein 2
|
|
chr14_+_102777433
Show fit
|
0.56 |
ENST00000392745.8
|
TRAF3
|
TNF receptor associated factor 3
|
|
chr12_+_6199715
Show fit
|
0.56 |
ENST00000382518.6
ENST00000642746.1
ENST00000538834.6
|
CD9
|
CD9 molecule
|
|
chr8_+_49911604
Show fit
|
0.55 |
ENST00000642164.1
ENST00000644093.1
ENST00000643999.1
ENST00000647073.1
ENST00000646880.1
|
SNTG1
|
syntrophin gamma 1
|
|
chr15_-_43302255
Show fit
|
0.55 |
ENST00000452443.3
|
TGM7
|
transglutaminase 7
|
|
chr11_+_64285219
Show fit
|
0.55 |
ENST00000377702.8
|
GPR137
|
G protein-coupled receptor 137
|
|
chr2_+_124025280
Show fit
|
0.55 |
ENST00000431078.1
ENST00000682447.1
|
CNTNAP5
|
contactin associated protein family member 5
|
|
chr20_-_44651683
Show fit
|
0.54 |
ENST00000537820.1
ENST00000372874.9
|
ADA
|
adenosine deaminase
|
|
chr10_-_102837406
Show fit
|
0.54 |
ENST00000369887.4
ENST00000638272.1
ENST00000639393.1
ENST00000638971.1
ENST00000638190.1
|
CYP17A1
|
cytochrome P450 family 17 subfamily A member 1
|
|
chrX_-_66033664
Show fit
|
0.54 |
ENST00000427538.5
|
VSIG4
|
V-set and immunoglobulin domain containing 4
|
|
chr6_+_132570322
Show fit
|
0.54 |
ENST00000275198.1
|
TAAR6
|
trace amine associated receptor 6
|
|
chr6_+_101393699
Show fit
|
0.54 |
ENST00000369134.9
ENST00000684068.1
ENST00000683903.1
ENST00000681975.1
|
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2
|
|
chr3_-_155854375
Show fit
|
0.54 |
ENST00000643144.2
ENST00000359479.7
ENST00000646424.1
|
SLC33A1
|
solute carrier family 33 member 1
|
|
chr7_+_104328603
Show fit
|
0.52 |
ENST00000401970.3
ENST00000424859.7
|
LHFPL3
|
LHFPL tetraspan subfamily member 3
|
|
chr16_-_66925526
Show fit
|
0.52 |
ENST00000299759.11
ENST00000420652.5
|
RRAD
|
RRAD, Ras related glycolysis inhibitor and calcium channel regulator
|
|
chr1_+_180196536
Show fit
|
0.52 |
ENST00000443059.1
|
QSOX1
|
quiescin sulfhydryl oxidase 1
|
|
chr2_+_218880844
Show fit
|
0.51 |
ENST00000258411.8
|
WNT10A
|
Wnt family member 10A
|
|
chr3_+_161221280
Show fit
|
0.50 |
ENST00000472947.5
ENST00000351193.7
ENST00000463518.5
|
NMD3
|
NMD3 ribosome export adaptor
|
|
chr7_-_138001794
Show fit
|
0.50 |
ENST00000616381.4
ENST00000620715.4
|
CREB3L2
|
cAMP responsive element binding protein 3 like 2
|
|
chr11_+_75717811
Show fit
|
0.49 |
ENST00000198801.10
|
MOGAT2
|
monoacylglycerol O-acyltransferase 2
|
|
chr3_+_184314525
Show fit
|
0.49 |
ENST00000392537.6
ENST00000444134.5
ENST00000450424.5
ENST00000421110.5
ENST00000435046.7
ENST00000382330.7
ENST00000426123.5
ENST00000346169.7
ENST00000350481.9
ENST00000455679.5
ENST00000440448.5
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma 1
|
|
chr8_+_49911396
Show fit
|
0.48 |
ENST00000642720.2
|
SNTG1
|
syntrophin gamma 1
|
|
chr11_+_64284982
Show fit
|
0.48 |
ENST00000539833.5
|
GPR137
|
G protein-coupled receptor 137
|
|
chr2_+_207711631
Show fit
|
0.47 |
ENST00000295414.8
ENST00000420822.1
ENST00000339882.9
|
CCNYL1
|
cyclin Y like 1
|
|
chr6_-_136526177
Show fit
|
0.47 |
ENST00000617204.4
|
MAP7
|
microtubule associated protein 7
|
|
chr4_+_61200318
Show fit
|
0.47 |
ENST00000683033.1
|
ADGRL3
|
adhesion G protein-coupled receptor L3
|
|
chr17_-_42676980
Show fit
|
0.46 |
ENST00000587627.1
ENST00000591022.6
ENST00000293349.10
|
PLEKHH3
|
pleckstrin homology, MyTH4 and FERM domain containing H3
|
|
chr1_+_23791203
Show fit
|
0.46 |
ENST00000374503.7
ENST00000374502.7
|
LYPLA2
|
lysophospholipase 2
|
|
chr2_-_89100352
Show fit
|
0.46 |
ENST00000479981.1
|
IGKV1-16
|
immunoglobulin kappa variable 1-16
|
|
chr1_-_74673786
Show fit
|
0.45 |
ENST00000326665.10
|
ERICH3
|
glutamate rich 3
|
|
chr12_-_8662619
Show fit
|
0.45 |
ENST00000544889.1
ENST00000543369.5
|
MFAP5
|
microfibril associated protein 5
|
|
chr11_+_64284822
Show fit
|
0.45 |
ENST00000536667.5
|
GPR137
|
G protein-coupled receptor 137
|
|
chr3_+_113947901
Show fit
|
0.45 |
ENST00000330212.7
ENST00000498275.5
|
ZDHHC23
|
zinc finger DHHC-type palmitoyltransferase 23
|
|
chr12_-_14567714
Show fit
|
0.45 |
ENST00000240617.10
|
PLBD1
|
phospholipase B domain containing 1
|
|
chr11_+_61792878
Show fit
|
0.44 |
ENST00000305885.3
ENST00000535723.1
|
FEN1
|
flap structure-specific endonuclease 1
|
|
chr3_+_133805789
Show fit
|
0.44 |
ENST00000678299.1
|
SRPRB
|
SRP receptor subunit beta
|
|
chr8_+_100158576
Show fit
|
0.44 |
ENST00000388798.7
|
SPAG1
|
sperm associated antigen 1
|
|
chr2_+_153871909
Show fit
|
0.43 |
ENST00000392825.8
ENST00000434213.1
|
GALNT13
|
polypeptide N-acetylgalactosaminyltransferase 13
|
|
chr20_-_2470749
Show fit
|
0.43 |
ENST00000381342.7
ENST00000438552.6
|
SNRPB
|
small nuclear ribonucleoprotein polypeptides B and B1
|
|
chr21_+_44455471
Show fit
|
0.43 |
ENST00000291592.6
|
LRRC3
|
leucine rich repeat containing 3
|
|
chr1_-_53738024
Show fit
|
0.42 |
ENST00000628545.1
|
GLIS1
|
GLIS family zinc finger 1
|
|
chr19_-_4540028
Show fit
|
0.42 |
ENST00000306390.7
|
LRG1
|
leucine rich alpha-2-glycoprotein 1
|
|
chr2_+_158968608
Show fit
|
0.41 |
ENST00000263635.8
|
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1
|
|
chr20_-_34303345
Show fit
|
0.41 |
ENST00000217426.7
|
AHCY
|
adenosylhomocysteinase
|
|
chr21_-_41953997
Show fit
|
0.41 |
ENST00000380486.4
|
C2CD2
|
C2 calcium dependent domain containing 2
|
|
chr6_+_36196710
Show fit
|
0.41 |
ENST00000357641.10
|
BRPF3
|
bromodomain and PHD finger containing 3
|
|
chr1_-_202159977
Show fit
|
0.41 |
ENST00000367279.8
|
PTPN7
|
protein tyrosine phosphatase non-receptor type 7
|
|
chr1_-_99005003
Show fit
|
0.41 |
ENST00000672681.1
ENST00000370188.7
|
PLPPR5
|
phospholipid phosphatase related 5
|
|
chr19_-_41363930
Show fit
|
0.41 |
ENST00000675972.1
|
B9D2
|
B9 domain containing 2
|
|
chr8_+_54616078
Show fit
|
0.40 |
ENST00000220676.2
|
RP1
|
RP1 axonemal microtubule associated
|
|
chr1_-_202161575
Show fit
|
0.40 |
ENST00000309017.7
ENST00000477554.5
ENST00000492451.1
|
PTPN7
|
protein tyrosine phosphatase non-receptor type 7
|
|
chr17_-_7238171
Show fit
|
0.40 |
ENST00000574236.5
ENST00000572789.5
|
PHF23
|
PHD finger protein 23
|
|
chr20_+_59300703
Show fit
|
0.40 |
ENST00000395654.3
|
EDN3
|
endothelin 3
|
|
chr1_+_37474572
Show fit
|
0.40 |
ENST00000373087.7
|
ZC3H12A
|
zinc finger CCCH-type containing 12A
|
|
chr16_+_280448
Show fit
|
0.40 |
ENST00000447871.5
|
ARHGDIG
|
Rho GDP dissociation inhibitor gamma
|
|
chr2_+_231781669
Show fit
|
0.40 |
ENST00000410024.5
ENST00000611614.4
ENST00000409295.5
ENST00000409091.5
|
COPS7B
|
COP9 signalosome subunit 7B
|
|
chr6_+_138161932
Show fit
|
0.39 |
ENST00000251691.5
|
ARFGEF3
|
ARFGEF family member 3
|
|
chr8_-_33567118
Show fit
|
0.39 |
ENST00000256257.2
|
RNF122
|
ring finger protein 122
|
|
chr8_+_22165358
Show fit
|
0.39 |
ENST00000306349.13
ENST00000306385.10
|
BMP1
|
bone morphogenetic protein 1
|
|
chr9_-_127930777
Show fit
|
0.39 |
ENST00000388747.9
|
PIP5KL1
|
phosphatidylinositol-4-phosphate 5-kinase like 1
|
|
chr18_+_36297661
Show fit
|
0.39 |
ENST00000257209.8
ENST00000590592.5
ENST00000359247.8
|
FHOD3
|
formin homology 2 domain containing 3
|
|
chr7_+_86643902
Show fit
|
0.38 |
ENST00000361669.7
|
GRM3
|
glutamate metabotropic receptor 3
|
|
chr10_+_68901258
Show fit
|
0.38 |
ENST00000373585.8
|
DDX50
|
DExD-box helicase 50
|
|
chr17_+_7380534
Show fit
|
0.38 |
ENST00000570896.5
|
TNK1
|
tyrosine kinase non receptor 1
|
|
chrX_+_103776493
Show fit
|
0.38 |
ENST00000433491.5
ENST00000612423.4
ENST00000443502.5
|
PLP1
|
proteolipid protein 1
|
|
chr3_+_184314495
Show fit
|
0.37 |
ENST00000352767.7
ENST00000414031.5
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma 1
|
|
chr17_-_42388360
Show fit
|
0.37 |
ENST00000678960.1
ENST00000404395.3
ENST00000389272.7
ENST00000677421.1
ENST00000585517.5
ENST00000264657.10
ENST00000678048.1
ENST00000678674.1
ENST00000678913.1
ENST00000678572.1
ENST00000678906.1
|
STAT3
|
signal transducer and activator of transcription 3
|
|
chr17_+_67377413
Show fit
|
0.37 |
ENST00000580974.5
|
PITPNC1
|
phosphatidylinositol transfer protein cytoplasmic 1
|
|
chr11_+_75240761
Show fit
|
0.37 |
ENST00000562197.3
|
TPBGL
|
trophoblast glycoprotein like
|
|
chr20_-_4815100
Show fit
|
0.36 |
ENST00000379376.2
|
RASSF2
|
Ras association domain family member 2
|
|
chr1_-_6261053
Show fit
|
0.36 |
ENST00000377893.3
|
GPR153
|
G protein-coupled receptor 153
|