|
chr5_+_132073782
Show fit
|
5.28 |
ENST00000296871.4
|
CSF2
|
colony stimulating factor 2
|
|
chr1_+_109910485
Show fit
|
2.57 |
ENST00000525659.5
|
CSF1
|
colony stimulating factor 1
|
|
chr21_-_35049238
Show fit
|
2.19 |
ENST00000416754.1
ENST00000437180.5
ENST00000455571.5
ENST00000675419.1
|
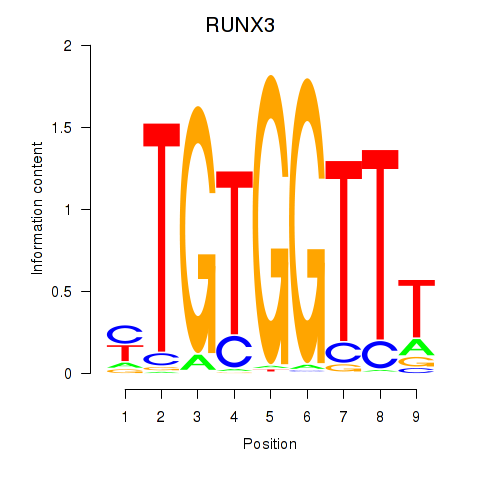
RUNX1
|
RUNX family transcription factor 1
|
|
chr21_-_35049327
Show fit
|
2.11 |
ENST00000300305.7
|
RUNX1
|
RUNX family transcription factor 1
|
|
chr11_-_128587551
Show fit
|
1.98 |
ENST00000392668.8
|
ETS1
|
ETS proto-oncogene 1, transcription factor
|
|
chr2_+_227813834
Show fit
|
1.93 |
ENST00000358813.5
ENST00000409189.7
|
CCL20
|
C-C motif chemokine ligand 20
|
|
chr5_+_35856883
Show fit
|
1.86 |
ENST00000506850.5
ENST00000303115.8
ENST00000511982.1
|
IL7R
|
interleukin 7 receptor
|
|
chr15_+_67125707
Show fit
|
1.84 |
ENST00000540846.6
|
SMAD3
|
SMAD family member 3
|
|
chr3_-_151203201
Show fit
|
1.81 |
ENST00000480322.1
ENST00000309180.6
|
GPR171
|
G protein-coupled receptor 171
|
|
chr1_+_109910840
Show fit
|
1.76 |
ENST00000329608.11
ENST00000488198.5
|
CSF1
|
colony stimulating factor 1
|
|
chr1_+_109910986
Show fit
|
1.67 |
ENST00000369801.1
|
CSF1
|
colony stimulating factor 1
|
|
chr2_+_102418642
Show fit
|
1.37 |
ENST00000264260.6
|
IL18RAP
|
interleukin 18 receptor accessory protein
|
|
chr6_+_45328203
Show fit
|
1.22 |
ENST00000371432.7
ENST00000647337.2
ENST00000371438.5
|
RUNX2
|
RUNX family transcription factor 2
|
|
chr1_-_173205543
Show fit
|
1.15 |
ENST00000367718.5
|
TNFSF4
|
TNF superfamily member 4
|
|
chrX_+_68829009
Show fit
|
1.01 |
ENST00000204961.5
|
EFNB1
|
ephrin B1
|
|
chr6_+_150721073
Show fit
|
1.00 |
ENST00000358517.6
|
PLEKHG1
|
pleckstrin homology and RhoGEF domain containing G1
|
|
chr17_-_63699730
Show fit
|
0.90 |
ENST00000578061.5
|
LIMD2
|
LIM domain containing 2
|
|
chr19_-_49155130
Show fit
|
0.90 |
ENST00000595625.1
|
HRC
|
histidine rich calcium binding protein
|
|
chr5_-_139482341
Show fit
|
0.77 |
ENST00000651699.1
|
STING1
|
stimulator of interferon response cGAMP interactor 1
|
|
chr12_+_121132869
Show fit
|
0.75 |
ENST00000328963.10
|
P2RX7
|
purinergic receptor P2X 7
|
|
chr5_-_139482285
Show fit
|
0.74 |
ENST00000652110.1
|
STING1
|
stimulator of interferon response cGAMP interactor 1
|
|
chr3_-_71130963
Show fit
|
0.73 |
ENST00000649695.2
|
FOXP1
|
forkhead box P1
|
|
chr3_-_71130557
Show fit
|
0.72 |
ENST00000497355.7
|
FOXP1
|
forkhead box P1
|
|
chr5_-_139482173
Show fit
|
0.71 |
ENST00000652271.1
|
STING1
|
stimulator of interferon response cGAMP interactor 1
|
|
chr1_-_173207322
Show fit
|
0.66 |
ENST00000281834.4
|
TNFSF4
|
TNF superfamily member 4
|
|
chr3_-_71130892
Show fit
|
0.66 |
ENST00000491238.7
ENST00000674446.1
|
FOXP1
|
forkhead box P1
|
|
chr19_+_10086787
Show fit
|
0.65 |
ENST00000590378.5
ENST00000397881.7
|
SHFL
|
shiftless antiviral inhibitor of ribosomal frameshifting
|
|
chr3_+_141386393
Show fit
|
0.65 |
ENST00000503809.5
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr19_-_49155384
Show fit
|
0.65 |
ENST00000252825.9
|
HRC
|
histidine rich calcium binding protein
|
|
chr3_-_99850976
Show fit
|
0.63 |
ENST00000487087.5
|
FILIP1L
|
filamin A interacting protein 1 like
|
|
chr12_+_75481204
Show fit
|
0.63 |
ENST00000550491.1
|
GLIPR1
|
GLI pathogenesis related 1
|
|
chr3_+_141386862
Show fit
|
0.61 |
ENST00000513258.5
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr5_+_134115563
Show fit
|
0.61 |
ENST00000517851.5
ENST00000521639.5
ENST00000522375.5
ENST00000378560.8
ENST00000520958.5
ENST00000518915.5
ENST00000395023.5
|
TCF7
|
transcription factor 7
|
|
chr5_-_95081482
Show fit
|
0.60 |
ENST00000312216.12
ENST00000512425.5
ENST00000505208.5
ENST00000429576.6
ENST00000508509.5
ENST00000510732.5
|
MCTP1
|
multiple C2 and transmembrane domain containing 1
|
|
chr14_+_51489112
Show fit
|
0.59 |
ENST00000356218.8
|
FRMD6
|
FERM domain containing 6
|
|
chr7_+_30852273
Show fit
|
0.57 |
ENST00000509504.2
|
ENSG00000250424.4
|
novel protein, MINDY4 and AQP1 readthrough
|
|
chr8_-_118951876
Show fit
|
0.53 |
ENST00000297350.9
|
TNFRSF11B
|
TNF receptor superfamily member 11b
|
|
chr17_-_58417521
Show fit
|
0.53 |
ENST00000584437.5
ENST00000407977.7
|
RNF43
|
ring finger protein 43
|
|
chr15_-_55270874
Show fit
|
0.53 |
ENST00000567380.5
ENST00000565972.5
ENST00000569493.5
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr7_-_116030735
Show fit
|
0.50 |
ENST00000393485.5
|
TFEC
|
transcription factor EC
|
|
chr3_-_190122317
Show fit
|
0.49 |
ENST00000427335.6
|
P3H2
|
prolyl 3-hydroxylase 2
|
|
chr17_-_58417547
Show fit
|
0.49 |
ENST00000577716.5
|
RNF43
|
ring finger protein 43
|
|
chr11_+_35176696
Show fit
|
0.49 |
ENST00000528455.5
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr9_+_108862255
Show fit
|
0.48 |
ENST00000333999.5
|
ACTL7A
|
actin like 7A
|
|
chr19_-_42132391
Show fit
|
0.47 |
ENST00000528894.8
ENST00000560804.6
ENST00000560558.5
ENST00000560398.5
ENST00000526816.6
ENST00000625670.2
|
POU2F2
|
POU class 2 homeobox 2
|
|
chr19_-_48993300
Show fit
|
0.47 |
ENST00000323798.8
ENST00000263276.6
|
GYS1
|
glycogen synthase 1
|
|
chr1_+_156369202
Show fit
|
0.46 |
ENST00000537040.6
|
RHBG
|
Rh family B glycoprotein
|
|
chr7_+_18495723
Show fit
|
0.45 |
ENST00000681950.1
ENST00000622668.4
ENST00000405010.7
ENST00000406451.8
ENST00000441542.7
ENST00000428307.6
ENST00000681273.1
|
HDAC9
|
histone deacetylase 9
|
|
chr11_+_35176575
Show fit
|
0.45 |
ENST00000526000.6
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr15_-_55270383
Show fit
|
0.45 |
ENST00000396307.6
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr15_-_55270280
Show fit
|
0.44 |
ENST00000564609.5
|
RAB27A
|
RAB27A, member RAS oncogene family
|
|
chr2_+_135741717
Show fit
|
0.44 |
ENST00000415164.5
|
UBXN4
|
UBX domain protein 4
|
|
chr10_+_99659430
Show fit
|
0.43 |
ENST00000370489.5
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7
|
|
chr9_+_116153783
Show fit
|
0.43 |
ENST00000328252.4
|
PAPPA
|
pappalysin 1
|
|
chr1_-_151826085
Show fit
|
0.43 |
ENST00000356728.11
|
RORC
|
RAR related orphan receptor C
|
|
chr19_-_42132465
Show fit
|
0.42 |
ENST00000529067.5
ENST00000529952.5
ENST00000342301.8
ENST00000389341.9
|
POU2F2
|
POU class 2 homeobox 2
|
|
chr6_+_31546845
Show fit
|
0.42 |
ENST00000376146.8
|
NFKBIL1
|
NFKB inhibitor like 1
|
|
chr17_+_82237134
Show fit
|
0.42 |
ENST00000583025.1
|
SLC16A3
|
solute carrier family 16 member 3
|
|
chr11_+_35176611
Show fit
|
0.41 |
ENST00000279452.10
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chr3_+_114294020
Show fit
|
0.41 |
ENST00000383671.8
|
TIGIT
|
T cell immunoreceptor with Ig and ITIM domains
|
|
chr9_-_34048868
Show fit
|
0.41 |
ENST00000379239.9
ENST00000684158.1
ENST00000379238.7
ENST00000360802.6
|
UBAP2
|
ubiquitin associated protein 2
|
|
chr11_+_35176639
Show fit
|
0.40 |
ENST00000527889.6
|
CD44
|
CD44 molecule (Indian blood group)
|
|
chrX_+_9463272
Show fit
|
0.39 |
ENST00000407597.7
ENST00000380961.5
ENST00000424279.6
|
TBL1X
|
transducin beta like 1 X-linked
|
|
chr3_-_11568764
Show fit
|
0.39 |
ENST00000424529.6
|
VGLL4
|
vestigial like family member 4
|
|
chr8_-_71361860
Show fit
|
0.39 |
ENST00000303824.11
ENST00000645451.1
|
EYA1
|
EYA transcriptional coactivator and phosphatase 1
|
|
chr17_+_20073538
Show fit
|
0.38 |
ENST00000681116.1
ENST00000680572.1
ENST00000680604.1
ENST00000681875.1
ENST00000679058.1
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1
|
|
chr12_+_14973020
Show fit
|
0.37 |
ENST00000266395.3
|
PDE6H
|
phosphodiesterase 6H
|
|
chr16_+_84768246
Show fit
|
0.36 |
ENST00000569925.1
ENST00000567526.1
|
USP10
|
ubiquitin specific peptidase 10
|
|
chr20_+_33079640
Show fit
|
0.36 |
ENST00000375483.4
|
BPIFB4
|
BPI fold containing family B member 4
|
|
chr15_+_73683938
Show fit
|
0.35 |
ENST00000567189.5
|
CD276
|
CD276 molecule
|
|
chr4_+_94489030
Show fit
|
0.35 |
ENST00000510099.5
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr8_-_71362054
Show fit
|
0.35 |
ENST00000340726.8
|
EYA1
|
EYA transcriptional coactivator and phosphatase 1
|
|
chr2_+_74529923
Show fit
|
0.35 |
ENST00000258080.8
ENST00000352222.7
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr15_-_43618800
Show fit
|
0.35 |
ENST00000450892.7
ENST00000541030.5
|
STRC
|
stereocilin
|
|
chr9_-_115091018
Show fit
|
0.34 |
ENST00000542877.5
ENST00000537320.5
ENST00000341037.8
|
TNC
|
tenascin C
|
|
chr1_+_239719542
Show fit
|
0.33 |
ENST00000448020.1
|
CHRM3
|
cholinergic receptor muscarinic 3
|
|
chr4_+_87975667
Show fit
|
0.33 |
ENST00000237623.11
ENST00000682655.1
ENST00000508233.6
ENST00000360804.4
ENST00000395080.8
|
SPP1
|
secreted phosphoprotein 1
|
|
chr2_+_96536743
Show fit
|
0.33 |
ENST00000673792.1
ENST00000357485.8
|
ARID5A
|
AT-rich interaction domain 5A
|
|
chr4_+_70226116
Show fit
|
0.33 |
ENST00000317987.6
|
FDCSP
|
follicular dendritic cell secreted protein
|
|
chr11_-_5324297
Show fit
|
0.33 |
ENST00000624187.1
|
OR51B2
|
olfactory receptor family 51 subfamily B member 2
|
|
chr20_-_46406559
Show fit
|
0.32 |
ENST00000372176.5
|
ELMO2
|
engulfment and cell motility 2
|
|
chr19_+_13024573
Show fit
|
0.32 |
ENST00000358552.7
ENST00000360105.8
ENST00000588228.5
ENST00000676441.1
ENST00000591028.1
|
NFIX
|
nuclear factor I X
|
|
chr1_-_217076889
Show fit
|
0.32 |
ENST00000493748.5
ENST00000463665.5
|
ESRRG
|
estrogen related receptor gamma
|
|
chr15_+_68290206
Show fit
|
0.31 |
ENST00000566008.1
|
FEM1B
|
fem-1 homolog B
|
|
chr1_-_161307420
Show fit
|
0.31 |
ENST00000491222.5
ENST00000672287.2
|
MPZ
|
myelin protein zero
|
|
chr10_-_13234368
Show fit
|
0.31 |
ENST00000378681.8
|
UCMA
|
upper zone of growth plate and cartilage matrix associated
|
|
chr20_-_46406582
Show fit
|
0.31 |
ENST00000450812.5
ENST00000290246.11
ENST00000396391.5
|
ELMO2
|
engulfment and cell motility 2
|
|
chr6_+_144583198
Show fit
|
0.30 |
ENST00000367526.8
|
UTRN
|
utrophin
|
|
chr4_+_87975829
Show fit
|
0.30 |
ENST00000614857.5
|
SPP1
|
secreted phosphoprotein 1
|
|
chr2_+_96537254
Show fit
|
0.29 |
ENST00000454558.2
|
ARID5A
|
AT-rich interaction domain 5A
|
|
chr10_+_25174969
Show fit
|
0.29 |
ENST00000376351.4
|
GPR158
|
G protein-coupled receptor 158
|
|
chr22_-_30246739
Show fit
|
0.29 |
ENST00000403987.3
ENST00000249075.4
|
LIF
|
LIF interleukin 6 family cytokine
|
|
chr7_+_120988683
Show fit
|
0.29 |
ENST00000340646.9
ENST00000310396.10
|
CPED1
|
cadherin like and PC-esterase domain containing 1
|
|
chr21_-_30372265
Show fit
|
0.29 |
ENST00000399889.4
|
KRTAP13-2
|
keratin associated protein 13-2
|
|
chr5_-_35938572
Show fit
|
0.29 |
ENST00000651391.1
ENST00000397366.5
ENST00000513623.5
ENST00000514524.2
ENST00000397367.6
|
CAPSL
|
calcyphosine like
|
|
chr8_-_133060347
Show fit
|
0.28 |
ENST00000427060.6
|
SLA
|
Src like adaptor
|
|
chr12_+_107685759
Show fit
|
0.28 |
ENST00000412830.8
ENST00000547995.5
|
PWP1
|
PWP1 homolog, endonuclein
|
|
chr2_+_135741814
Show fit
|
0.28 |
ENST00000272638.14
|
UBXN4
|
UBX domain protein 4
|
|
chr10_-_13234329
Show fit
|
0.28 |
ENST00000463405.2
|
UCMA
|
upper zone of growth plate and cartilage matrix associated
|
|
chr3_-_196287649
Show fit
|
0.27 |
ENST00000441879.5
ENST00000292823.6
ENST00000431016.6
ENST00000411591.5
|
PCYT1A
|
phosphate cytidylyltransferase 1, choline, alpha
|
|
chr7_-_116030750
Show fit
|
0.27 |
ENST00000265440.12
ENST00000320239.11
|
TFEC
|
transcription factor EC
|
|
chr5_+_136160986
Show fit
|
0.27 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5
|
|
chr18_-_58629084
Show fit
|
0.27 |
ENST00000361673.4
|
ALPK2
|
alpha kinase 2
|
|
chr1_-_113871665
Show fit
|
0.27 |
ENST00000528414.5
ENST00000460620.5
ENST00000359785.10
ENST00000420377.6
ENST00000525799.1
ENST00000538253.5
|
PTPN22
|
protein tyrosine phosphatase non-receptor type 22
|
|
chr11_-_66958366
Show fit
|
0.27 |
ENST00000651036.1
ENST00000652125.1
ENST00000531614.6
ENST00000524491.6
ENST00000529047.6
ENST00000393960.7
ENST00000393958.7
ENST00000528403.6
ENST00000651854.1
|
PC
|
pyruvate carboxylase
|
|
chr6_+_110874775
Show fit
|
0.27 |
ENST00000675380.1
ENST00000368882.8
ENST00000368877.9
ENST00000368885.8
ENST00000672937.2
|
AMD1
|
adenosylmethionine decarboxylase 1
|
|
chr1_-_168543990
Show fit
|
0.27 |
ENST00000367819.3
|
XCL2
|
X-C motif chemokine ligand 2
|
|
chr6_+_31575557
Show fit
|
0.27 |
ENST00000449264.3
|
TNF
|
tumor necrosis factor
|
|
chr17_+_7336502
Show fit
|
0.27 |
ENST00000158762.8
|
ACAP1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1
|
|
chr16_-_375205
Show fit
|
0.26 |
ENST00000448854.1
|
PGAP6
|
post-glycosylphosphatidylinositol attachment to proteins 6
|
|
chr15_-_89211803
Show fit
|
0.26 |
ENST00000563254.1
|
RLBP1
|
retinaldehyde binding protein 1
|
|
chr1_+_109910892
Show fit
|
0.26 |
ENST00000369802.7
ENST00000420111.6
|
CSF1
|
colony stimulating factor 1
|
|
chr2_+_238426920
Show fit
|
0.26 |
ENST00000264607.9
|
ASB1
|
ankyrin repeat and SOCS box containing 1
|
|
chrX_-_49184789
Show fit
|
0.26 |
ENST00000453382.5
ENST00000432913.5
|
PRICKLE3
|
prickle planar cell polarity protein 3
|
|
chr1_+_209686173
Show fit
|
0.26 |
ENST00000615289.4
ENST00000367028.6
ENST00000261465.5
|
HSD11B1
|
hydroxysteroid 11-beta dehydrogenase 1
|
|
chr16_-_67964044
Show fit
|
0.26 |
ENST00000537830.6
|
SLC12A4
|
solute carrier family 12 member 4
|
|
chr1_+_50103903
Show fit
|
0.25 |
ENST00000371827.5
|
ELAVL4
|
ELAV like RNA binding protein 4
|
|
chr3_+_141324208
Show fit
|
0.25 |
ENST00000509842.5
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr1_-_85578345
Show fit
|
0.25 |
ENST00000426972.8
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr2_+_74530018
Show fit
|
0.25 |
ENST00000437202.1
|
HTRA2
|
HtrA serine peptidase 2
|
|
chr10_+_25174796
Show fit
|
0.25 |
ENST00000650135.1
|
GPR158
|
G protein-coupled receptor 158
|
|
chr20_+_44714853
Show fit
|
0.25 |
ENST00000372865.4
|
CCN5
|
cellular communication network factor 5
|
|
chr5_-_159208066
Show fit
|
0.25 |
ENST00000519865.5
ENST00000521606.6
|
RNF145
|
ring finger protein 145
|
|
chr6_+_116370938
Show fit
|
0.24 |
ENST00000644252.3
ENST00000646710.1
ENST00000359564.3
|
DSE
|
dermatan sulfate epimerase
|
|
chrX_-_71618455
Show fit
|
0.24 |
ENST00000373691.4
ENST00000373693.4
|
CXCR3
|
C-X-C motif chemokine receptor 3
|
|
chr9_-_120877026
Show fit
|
0.24 |
ENST00000436309.5
|
PHF19
|
PHD finger protein 19
|
|
chr19_-_43780957
Show fit
|
0.24 |
ENST00000648319.1
|
KCNN4
|
potassium calcium-activated channel subfamily N member 4
|
|
chr9_-_120876356
Show fit
|
0.24 |
ENST00000456291.1
|
PHF19
|
PHD finger protein 19
|
|
chr2_-_49154433
Show fit
|
0.24 |
ENST00000454032.5
ENST00000304421.8
|
FSHR
|
follicle stimulating hormone receptor
|
|
chr15_+_73684204
Show fit
|
0.24 |
ENST00000537340.6
ENST00000318424.9
|
CD276
|
CD276 molecule
|
|
chr20_+_44714835
Show fit
|
0.23 |
ENST00000372868.6
|
CCN5
|
cellular communication network factor 5
|
|
chr10_+_122374685
Show fit
|
0.23 |
ENST00000368990.7
ENST00000368989.6
ENST00000463663.6
|
PLEKHA1
|
pleckstrin homology domain containing A1
|
|
chr2_-_49154507
Show fit
|
0.23 |
ENST00000406846.7
|
FSHR
|
follicle stimulating hormone receptor
|
|
chr1_+_209768597
Show fit
|
0.23 |
ENST00000487271.5
ENST00000477431.1
|
TRAF3IP3
|
TRAF3 interacting protein 3
|
|
chr13_-_102401599
Show fit
|
0.23 |
ENST00000376131.8
|
FGF14
|
fibroblast growth factor 14
|
|
chr2_-_18560616
Show fit
|
0.23 |
ENST00000381249.4
|
RDH14
|
retinol dehydrogenase 14
|
|
chr1_-_94925759
Show fit
|
0.23 |
ENST00000415017.1
ENST00000545882.5
|
CNN3
|
calponin 3
|
|
chr11_-_31803620
Show fit
|
0.22 |
ENST00000639006.1
|
PAX6
|
paired box 6
|
|
chr22_-_45240859
Show fit
|
0.22 |
ENST00000336156.10
|
KIAA0930
|
KIAA0930
|
|
chrX_-_15854743
Show fit
|
0.22 |
ENST00000450644.2
|
AP1S2
|
adaptor related protein complex 1 subunit sigma 2
|
|
chr2_+_26848199
Show fit
|
0.22 |
ENST00000401478.5
|
DPYSL5
|
dihydropyrimidinase like 5
|
|
chr6_+_31547560
Show fit
|
0.22 |
ENST00000376148.9
ENST00000376145.8
|
NFKBIL1
|
NFKB inhibitor like 1
|
|
chr3_+_98531965
Show fit
|
0.22 |
ENST00000284311.5
|
GPR15
|
G protein-coupled receptor 15
|
|
chr4_-_10684749
Show fit
|
0.22 |
ENST00000226951.11
|
CLNK
|
cytokine dependent hematopoietic cell linker
|
|
chr1_+_156338619
Show fit
|
0.22 |
ENST00000481479.5
ENST00000368252.5
ENST00000466306.5
ENST00000368251.1
|
TSACC
|
TSSK6 activating cochaperone
|
|
chr4_-_54064586
Show fit
|
0.21 |
ENST00000263921.8
|
CHIC2
|
cysteine rich hydrophobic domain 2
|
|
chr6_-_114343012
Show fit
|
0.21 |
ENST00000312719.10
|
HS3ST5
|
heparan sulfate-glucosamine 3-sulfotransferase 5
|
|
chr1_-_172444055
Show fit
|
0.21 |
ENST00000344529.5
ENST00000367728.1
|
PIGC
|
phosphatidylinositol glycan anchor biosynthesis class C
|
|
chr17_+_19378476
Show fit
|
0.20 |
ENST00000395604.8
ENST00000482850.1
|
MAPK7
|
mitogen-activated protein kinase 7
|
|
chr17_+_47733228
Show fit
|
0.20 |
ENST00000177694.2
|
TBX21
|
T-box transcription factor 21
|
|
chr22_+_21642287
Show fit
|
0.20 |
ENST00000248958.5
|
SDF2L1
|
stromal cell derived factor 2 like 1
|
|
chr19_-_50365625
Show fit
|
0.20 |
ENST00000598915.5
ENST00000253719.7
|
NAPSA
|
napsin A aspartic peptidase
|
|
chr1_+_209756149
Show fit
|
0.20 |
ENST00000367026.7
|
TRAF3IP3
|
TRAF3 interacting protein 3
|
|
chr16_+_88382950
Show fit
|
0.19 |
ENST00000565624.3
|
ZNF469
|
zinc finger protein 469
|
|
chr17_+_42844573
Show fit
|
0.19 |
ENST00000253799.8
ENST00000452774.2
|
AOC2
|
amine oxidase copper containing 2
|
|
chr17_+_81977587
Show fit
|
0.19 |
ENST00000306739.9
ENST00000581647.5
ENST00000580534.5
ENST00000579684.5
|
ASPSCR1
|
ASPSCR1 tether for SLC2A4, UBX domain containing
|
|
chr8_-_95269190
Show fit
|
0.19 |
ENST00000286688.6
|
C8orf37
|
chromosome 8 open reading frame 37
|
|
chr3_+_50246888
Show fit
|
0.19 |
ENST00000451956.1
|
GNAI2
|
G protein subunit alpha i2
|
|
chr11_+_71538025
Show fit
|
0.18 |
ENST00000398534.4
|
KRTAP5-8
|
keratin associated protein 5-8
|
|
chr1_-_206772484
Show fit
|
0.18 |
ENST00000423557.1
|
IL10
|
interleukin 10
|
|
chr1_+_172659095
Show fit
|
0.18 |
ENST00000367721.3
ENST00000340030.4
|
FASLG
|
Fas ligand
|
|
chr8_+_30387064
Show fit
|
0.18 |
ENST00000523115.5
ENST00000519647.5
|
RBPMS
|
RNA binding protein, mRNA processing factor
|
|
chr5_+_172959511
Show fit
|
0.18 |
ENST00000519522.1
|
RPL26L1
|
ribosomal protein L26 like 1
|
|
chr1_-_1779976
Show fit
|
0.18 |
ENST00000378625.5
|
NADK
|
NAD kinase
|
|
chr1_+_156338455
Show fit
|
0.18 |
ENST00000368253.6
ENST00000470342.5
ENST00000368254.6
|
TSACC
|
TSSK6 activating cochaperone
|
|
chr2_-_2324323
Show fit
|
0.18 |
ENST00000648339.1
ENST00000647694.1
|
MYT1L
|
myelin transcription factor 1 like
|
|
chr11_+_71548420
Show fit
|
0.18 |
ENST00000528743.4
|
KRTAP5-9
|
keratin associated protein 5-9
|
|
chr15_+_49423233
Show fit
|
0.18 |
ENST00000560270.1
ENST00000267843.9
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7
|
|
chr14_+_22883220
Show fit
|
0.18 |
ENST00000536884.1
ENST00000267396.9
|
REM2
|
RRAD and GEM like GTPase 2
|
|
chr12_-_16606795
Show fit
|
0.17 |
ENST00000447609.5
|
LMO3
|
LIM domain only 3
|
|
chr17_+_81977539
Show fit
|
0.17 |
ENST00000306729.11
|
ASPSCR1
|
ASPSCR1 tether for SLC2A4, UBX domain containing
|
|
chr1_+_181483510
Show fit
|
0.17 |
ENST00000367573.7
|
CACNA1E
|
calcium voltage-gated channel subunit alpha1 E
|
|
chr5_-_138543198
Show fit
|
0.17 |
ENST00000507939.5
ENST00000572514.5
ENST00000499810.6
ENST00000360541.10
|
ETF1
|
eukaryotic translation termination factor 1
|
|
chr12_+_123973197
Show fit
|
0.17 |
ENST00000392404.7
ENST00000337815.9
ENST00000538932.6
ENST00000618862.2
ENST00000389727.8
|
ZNF664
ENSG00000274874.2
RFLNA
|
zinc finger protein 664
novel protein
refilin A
|
|
chr17_+_19377721
Show fit
|
0.17 |
ENST00000308406.9
ENST00000299612.11
|
MAPK7
|
mitogen-activated protein kinase 7
|
|
chr16_-_11915878
Show fit
|
0.17 |
ENST00000439887.6
ENST00000434724.7
|
GSPT1
|
G1 to S phase transition 1
|
|
chr12_+_93378625
Show fit
|
0.17 |
ENST00000546925.1
|
NUDT4
|
nudix hydrolase 4
|
|
chr16_-_11915991
Show fit
|
0.17 |
ENST00000420576.6
|
GSPT1
|
G1 to S phase transition 1
|
|
chr3_-_33645433
Show fit
|
0.17 |
ENST00000635664.1
ENST00000485378.6
ENST00000313350.10
ENST00000487200.5
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr3_+_41199462
Show fit
|
0.16 |
ENST00000647264.1
ENST00000645900.1
ENST00000646174.1
ENST00000643992.1
ENST00000647390.1
ENST00000642886.1
|
CTNNB1
|
catenin beta 1
|
|
chr6_-_33289189
Show fit
|
0.16 |
ENST00000374617.9
|
WDR46
|
WD repeat domain 46
|
|
chr22_+_39901075
Show fit
|
0.16 |
ENST00000344138.9
|
GRAP2
|
GRB2 related adaptor protein 2
|
|
chr16_-_68448491
Show fit
|
0.16 |
ENST00000561749.1
ENST00000219334.10
|
SMPD3
|
sphingomyelin phosphodiesterase 3
|
|
chr10_+_24449426
Show fit
|
0.16 |
ENST00000307544.10
|
KIAA1217
|
KIAA1217
|
|
chr6_-_75202792
Show fit
|
0.16 |
ENST00000416123.6
|
COL12A1
|
collagen type XII alpha 1 chain
|
|
chr14_+_21768482
Show fit
|
0.16 |
ENST00000390428.3
|
TRAV6
|
T cell receptor alpha variable 6
|
|
chr7_-_29195186
Show fit
|
0.16 |
ENST00000449801.5
ENST00000409850.5
|
CPVL
|
carboxypeptidase vitellogenic like
|
|
chr19_-_15449920
Show fit
|
0.15 |
ENST00000263381.12
ENST00000643092.1
ENST00000673675.1
|
WIZ
|
WIZ zinc finger
|
|
chr2_+_167135901
Show fit
|
0.15 |
ENST00000628543.2
|
XIRP2
|
xin actin binding repeat containing 2
|
|
chr1_-_153544997
Show fit
|
0.15 |
ENST00000368715.5
|
S100A4
|
S100 calcium binding protein A4
|
|
chr14_+_35292429
Show fit
|
0.15 |
ENST00000555764.5
ENST00000556506.1
|
PSMA6
|
proteasome 20S subunit alpha 6
|
|
chr11_-_31803663
Show fit
|
0.15 |
ENST00000638802.1
ENST00000638878.1
|
PAX6
|
paired box 6
|
|
chr16_-_69384747
Show fit
|
0.14 |
ENST00000566257.5
|
TERF2
|
telomeric repeat binding factor 2
|
|
chr11_+_60971777
Show fit
|
0.14 |
ENST00000542157.5
ENST00000433107.6
ENST00000352009.9
ENST00000452451.6
|
CD6
|
CD6 molecule
|
|
chr2_-_213151590
Show fit
|
0.14 |
ENST00000374319.8
ENST00000457361.5
ENST00000451136.6
ENST00000434687.6
|
IKZF2
|
IKAROS family zinc finger 2
|
|
chr3_+_155083889
Show fit
|
0.14 |
ENST00000680282.1
|
MME
|
membrane metalloendopeptidase
|
|
chr16_+_85908988
Show fit
|
0.14 |
ENST00000566369.1
|
IRF8
|
interferon regulatory factor 8
|
|
chr3_+_155083523
Show fit
|
0.14 |
ENST00000680057.1
|
MME
|
membrane metalloendopeptidase
|
|
chr14_+_60509138
Show fit
|
0.13 |
ENST00000327720.6
|
SIX6
|
SIX homeobox 6
|
|
chr17_+_49210699
Show fit
|
0.13 |
ENST00000225941.6
|
ABI3
|
ABI family member 3
|