Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
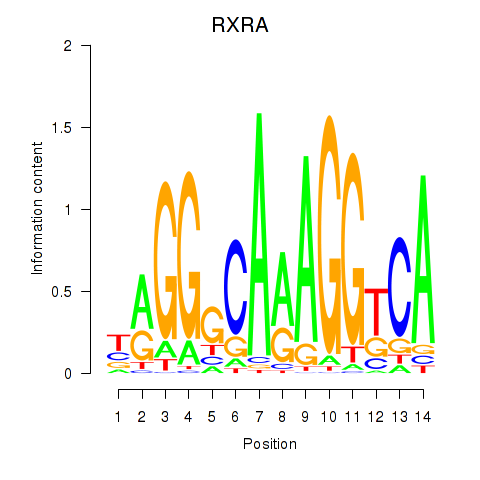
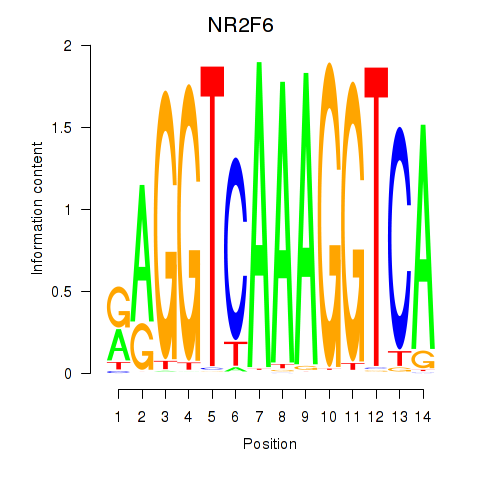
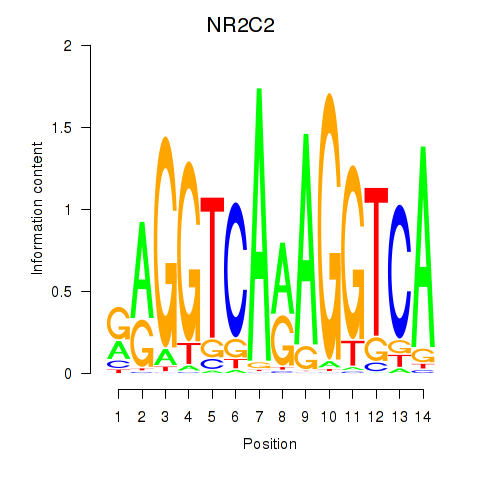
Results for RXRA_NR2F6_NR2C2
Z-value: 0.88
Transcription factors associated with RXRA_NR2F6_NR2C2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RXRA
|
ENSG00000186350.12 | RXRA |
|
NR2F6
|
ENSG00000160113.6 | NR2F6 |
|
NR2C2
|
ENSG00000177463.15 | NR2C2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2C2 | hg38_v1_chr3_+_14947568_14947606, hg38_v1_chr3_+_14947680_14947729 | -0.57 | 2.7e-03 | Click! |
| NR2F6 | hg38_v1_chr19_-_17245889_17245954 | -0.44 | 3.0e-02 | Click! |
| RXRA | hg38_v1_chr9_+_134326435_134326494 | 0.12 | 5.6e-01 | Click! |
Activity profile of RXRA_NR2F6_NR2C2 motif
Sorted Z-values of RXRA_NR2F6_NR2C2 motif
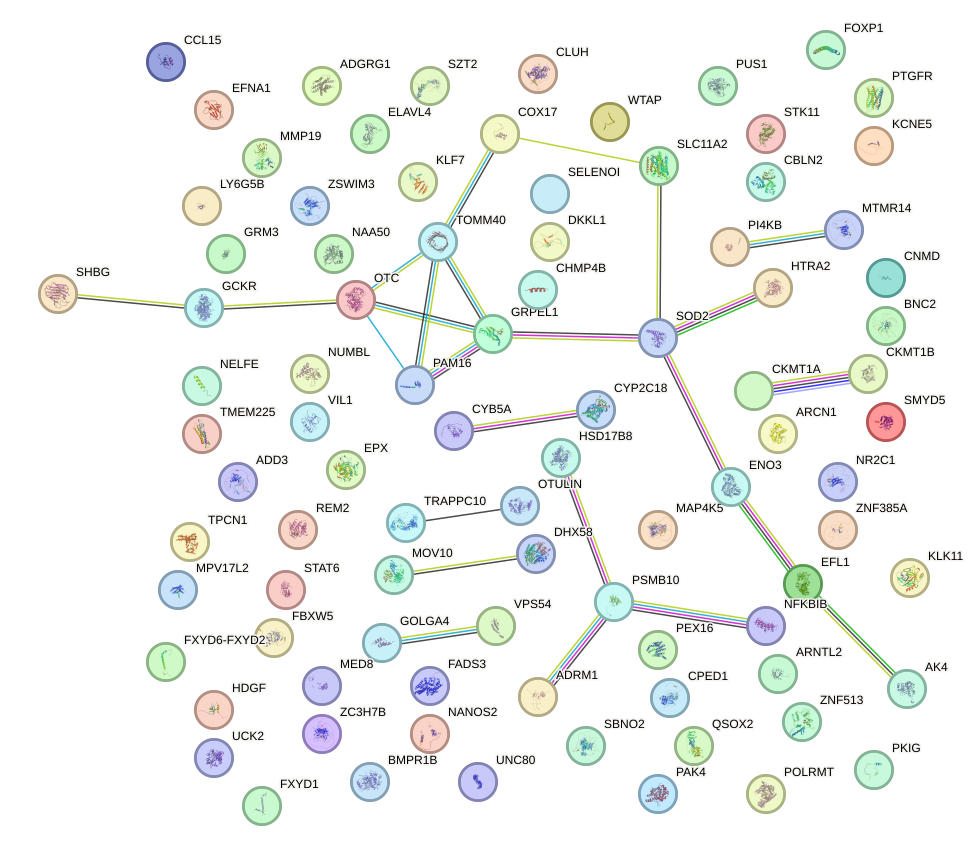
Network of associatons between targets according to the STRING database.
First level regulatory network of RXRA_NR2F6_NR2C2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.4 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 1.6 | 7.8 | GO:0003070 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 1.4 | 7.1 | GO:2000327 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.6 | 2.5 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.5 | 6.1 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.4 | 1.2 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.4 | 1.5 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.4 | 3.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.3 | 1.3 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.3 | 1.3 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.3 | 0.9 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.3 | 1.7 | GO:0010731 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.3 | 1.6 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.2 | 2.4 | GO:0001554 | luteolysis(GO:0001554) |
| 0.2 | 2.5 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.2 | 0.9 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.2 | 0.7 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.2 | 0.9 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.2 | 1.0 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.2 | 3.8 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.2 | 0.6 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.2 | 0.7 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.2 | 1.3 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.2 | 0.7 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.2 | 0.9 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.2 | 0.5 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.2 | 1.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 0.4 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.1 | 0.5 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 0.5 | GO:0060327 | cytoplasmic actin-based contraction involved in cell motility(GO:0060327) |
| 0.1 | 0.5 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.4 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.1 | 1.0 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.4 | GO:0014810 | positive regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014810) |
| 0.1 | 0.3 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 0.1 | 0.6 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.1 | 1.0 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 1.0 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.1 | 0.3 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 | 0.7 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.1 | 1.7 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.4 | GO:1904732 | regulation of electron carrier activity(GO:1904732) |
| 0.1 | 0.6 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 0.3 | GO:0035565 | regulation of pronephros size(GO:0035565) |
| 0.1 | 1.6 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 1.7 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 0.3 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.1 | 1.4 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 0.2 | GO:0007174 | epidermal growth factor catabolic process(GO:0007174) |
| 0.1 | 1.3 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.1 | 0.4 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.7 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 1.2 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.5 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.1 | 0.2 | GO:1902724 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) positive regulation of skeletal muscle cell proliferation(GO:0014858) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.1 | 0.5 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.8 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 1.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 1.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.5 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 0.2 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 0.6 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.5 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.2 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.3 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.2 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 1.4 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.6 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.2 | GO:0046882 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.4 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.3 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.4 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.5 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 0.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.4 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.1 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.1 | GO:0001796 | type IIa hypersensitivity(GO:0001794) regulation of type IIa hypersensitivity(GO:0001796) type II hypersensitivity(GO:0002445) regulation of type II hypersensitivity(GO:0002892) |
| 0.0 | 0.3 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 1.3 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.4 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.4 | GO:0015684 | ferrous iron transport(GO:0015684) ferrous iron transmembrane transport(GO:1903874) |
| 0.0 | 0.1 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.3 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.9 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.2 | GO:1902943 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) regulation of voltage-gated chloride channel activity(GO:1902941) positive regulation of voltage-gated chloride channel activity(GO:1902943) positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.0 | 0.1 | GO:0036404 | conversion of ds siRNA to ss siRNA involved in RNA interference(GO:0033168) conversion of ds siRNA to ss siRNA(GO:0036404) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.5 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.6 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.2 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.6 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.3 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.2 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 1.2 | GO:0006735 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.2 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.4 | GO:1903543 | positive regulation of exosomal secretion(GO:1903543) |
| 0.0 | 0.7 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0052553 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 0.0 | 0.4 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.3 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.3 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.3 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.2 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.1 | GO:0036116 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.1 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.2 | GO:0060353 | regulation of cell adhesion molecule production(GO:0060353) |
| 0.0 | 0.4 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 1.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:1990927 | calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 1.6 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.2 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.2 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.2 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.1 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.0 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.0 | 0.1 | GO:0050915 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.4 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.4 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.6 | GO:0038202 | TORC1 signaling(GO:0038202) |
| 0.0 | 1.1 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 3.0 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.2 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.2 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 1.4 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.2 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 1.1 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 0.2 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.0 | GO:0002351 | serotonin production involved in inflammatory response(GO:0002351) serotonin secretion involved in inflammatory response(GO:0002442) serotonin secretion by platelet(GO:0002554) |
| 0.0 | 0.4 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.0 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.3 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.2 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.0 | GO:0014873 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) regulation of amino acid import(GO:0010958) response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 1.3 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.2 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.5 | 7.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.4 | 2.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.2 | 1.7 | GO:0032021 | NELF complex(GO:0032021) |
| 0.2 | 1.0 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.2 | 1.0 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.2 | 1.6 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.2 | 1.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.2 | 1.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 2.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.4 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.1 | 1.0 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 1.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.9 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.7 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 1.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.8 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 1.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.2 | GO:0032419 | extrinsic component of lysosome membrane(GO:0032419) |
| 0.1 | 0.9 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.4 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.3 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.1 | 0.2 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 0.6 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.3 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.3 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 1.7 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 1.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0033167 | ARC complex(GO:0033167) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 21.8 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.3 | GO:0000125 | PCAF complex(GO:0000125) pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.9 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 1.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 1.8 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.3 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.4 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 1.2 | 6.0 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.8 | 8.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.4 | 1.3 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.3 | 2.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.3 | 0.9 | GO:0052858 | peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) |
| 0.3 | 1.6 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.2 | 0.7 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.2 | 0.9 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.2 | 1.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.2 | 1.0 | GO:0015087 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.2 | 0.6 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.2 | 1.6 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 0.5 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.1 | 1.0 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 0.4 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.1 | 0.5 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 1.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.6 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 1.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.4 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.3 | GO:0090422 | thiamine pyrophosphate transporter activity(GO:0090422) |
| 0.1 | 0.5 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 1.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.9 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.5 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.1 | 0.7 | GO:0005497 | androgen binding(GO:0005497) |
| 0.1 | 1.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 6.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.4 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.1 | 0.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 1.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.1 | 1.9 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.3 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.3 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.1 | 0.6 | GO:0030613 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.1 | 0.4 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.1 | 0.4 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 0.5 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.3 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.1 | 0.5 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.5 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 1.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.2 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.2 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.0 | 0.3 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.2 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.3 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.9 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 3.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.0 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.2 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 1.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 1.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 1.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 1.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.6 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.4 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0047783 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.0 | 0.3 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.5 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 1.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 3.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.2 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.1 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.3 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 2.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 1.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.3 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 1.2 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.4 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.1 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 1.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.0 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.6 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 2.2 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 1.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 1.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.7 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.5 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.9 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.5 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.2 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.4 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.1 | 2.0 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 4.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.0 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 0.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 0.7 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 3.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.9 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 1.7 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 1.0 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 1.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.0 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 1.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 2.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.9 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 1.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME INFLUENZA LIFE CYCLE | Genes involved in Influenza Life Cycle |
| 0.0 | 0.5 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.3 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.1 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.6 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.7 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.0 | 0.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.1 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |