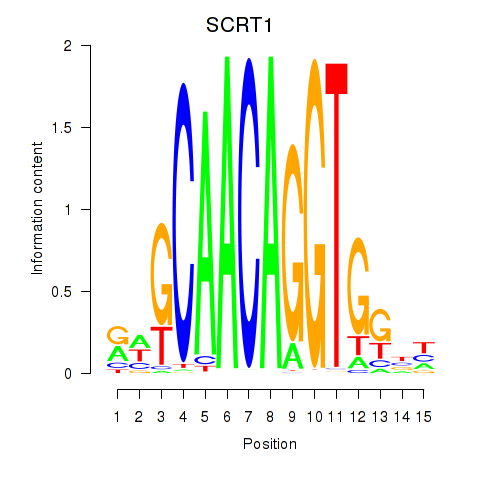
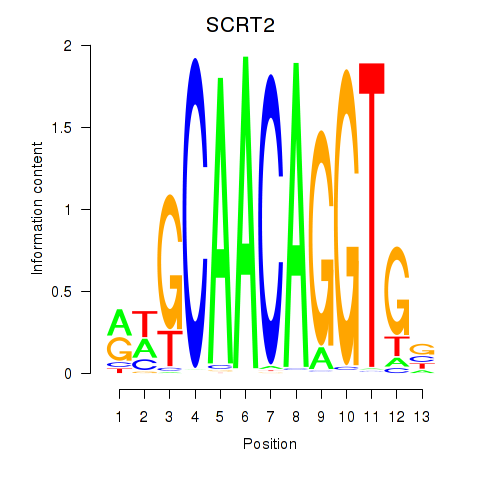
chr4_+_41538143
Show fit
8.27
ENST00000503057.6
ENST00000511496.5
LIMCH1
LIM and calponin homology domains 1
chr8_-_126557691
Show fit
4.36
ENST00000652209.1
LRATD2
LRAT domain containing 2
chr4_-_89836213
Show fit
3.99
ENST00000618500.4
ENST00000508895.5
SNCA
synuclein alpha
chr4_-_148442342
Show fit
3.77
ENST00000358102.8
NR3C2
nuclear receptor subfamily 3 group C member 2
chr4_-_148442508
Show fit
3.64
ENST00000625323.2
NR3C2
nuclear receptor subfamily 3 group C member 2
chr10_+_35195843
Show fit
2.75
ENST00000488741.5
ENST00000474931.5
ENST00000468236.5
ENST00000344351.5
ENST00000490511.1
CREM
cAMP responsive element modulator
chr11_+_59787067
Show fit
2.15
ENST00000528805.1
STX3
syntaxin 3
chr10_+_35195124
Show fit
1.96
ENST00000487763.5
ENST00000473940.5
ENST00000488328.5
ENST00000356917.9
CREM
cAMP responsive element modulator
chr7_-_28958321
Show fit
1.88
ENST00000539664.3
TRIL
TLR4 interactor with leucine rich repeats
chr5_-_74640575
Show fit
1.87
ENST00000651128.1
ENC1
ectodermal-neural cortex 1
chr5_-_74640719
Show fit
1.81
ENST00000302351.9
ENST00000509127.2
ENC1
ectodermal-neural cortex 1
chr3_+_16174628
Show fit
1.80
ENST00000339732.10
GALNT15
polypeptide N-acetylgalactosaminyltransferase 15
chr10_+_74824919
Show fit
1.75
ENST00000648828.1
ENST00000648725.1
KAT6B
lysine acetyltransferase 6B
chr5_-_74640649
Show fit
1.72
ENST00000537006.1
ENC1
ectodermal-neural cortex 1
chr1_+_15756659
Show fit
1.71
ENST00000375771.5
FBLIM1
filamin binding LIM protein 1
chr8_+_97869040
Show fit
1.68
ENST00000254898.7
ENST00000524308.5
ENST00000522025.6
MATN2
matrilin 2
chr19_-_11262499
Show fit
1.54
ENST00000294618.12
DOCK6
dedicator of cytokinesis 6
chr17_+_70169516
Show fit
1.53
ENST00000243457.4
KCNJ2
potassium inwardly rectifying channel subfamily J member 2
chr22_-_21735776
Show fit
1.49
ENST00000339468.8
YPEL1
yippee like 1
chr7_+_12687625
Show fit
1.47
ENST00000651779.1
ENST00000404894.1
ARL4A
ADP ribosylation factor like GTPase 4A
chr2_+_172735838
Show fit
1.42
ENST00000397081.8
RAPGEF4
Rap guanine nucleotide exchange factor 4
chr2_+_188291994
Show fit
1.41
ENST00000409927.5
ENST00000409805.5
GULP1
GULP PTB domain containing engulfment adaptor 1
chr6_+_15248855
Show fit
1.34
ENST00000397311.4
JARID2
jumonji and AT-rich interaction domain containing 2
chr8_+_119208322
Show fit
1.32
ENST00000614891.5
MAL2
mal, T cell differentiation protein 2
chrX_+_72069659
Show fit
1.31
ENST00000631375.1
NHSL2
NHS like 2
chr2_+_172735912
Show fit
1.31
ENST00000409036.5
RAPGEF4
Rap guanine nucleotide exchange factor 4
chr12_-_70609788
Show fit
1.28
ENST00000547715.1
ENST00000538708.5
ENST00000550857.5
ENST00000261266.9
PTPRB
protein tyrosine phosphatase receptor type B
chr2_+_188291854
Show fit
1.24
ENST00000409830.6
GULP1
GULP PTB domain containing engulfment adaptor 1
chr22_-_21735744
Show fit
1.24
ENST00000403503.1
YPEL1
yippee like 1
chr1_-_93847150
Show fit
1.21
ENST00000370244.5
BCAR3
BCAR3 adaptor protein, NSP family member
chrX_+_55717796
Show fit
1.20
ENST00000262850.7
RRAGB
Ras related GTP binding B
chrX_-_55030970
Show fit
1.20
ENST00000493869.2
ENST00000396198.7
ENST00000650242.1
ENST00000335854.8
ENST00000477869.6
ENST00000455688.2
ENST00000644983.1
ALAS2
5'-aminolevulinate synthase 2
chr9_+_131096476
Show fit
1.19
ENST00000372309.7
ENST00000247291.8
ENST00000372302.5
ENST00000372300.5
ENST00000372298.1
AIF1L
allograft inflammatory factor 1 like
chr5_-_177996242
Show fit
1.18
ENST00000308304.2
PROP1
PROP paired-like homeobox 1
chr5_-_111756245
Show fit
1.17
ENST00000447165.6
NREP
neuronal regeneration related protein
chr5_+_73626158
Show fit
1.15
ENST00000296794.10
ENST00000545377.5
ENST00000509848.5
ENST00000513042.7
ARHGEF28
Rho guanine nucleotide exchange factor 28
chr9_+_71911468
Show fit
1.11
ENST00000377031.7
C9orf85
chromosome 9 open reading frame 85
chr9_+_122941003
Show fit
1.09
ENST00000373647.9
ENST00000402311.5
RABGAP1
RAB GTPase activating protein 1
chr12_-_30695852
Show fit
1.03
ENST00000256079.9
IPO8
importin 8
chr15_-_29821473
Show fit
1.01
ENST00000400011.6
TJP1
tight junction protein 1
chr7_-_122886706
Show fit
1.01
ENST00000313070.11
ENST00000334010.11
ENST00000615869.4
CADPS2
calcium dependent secretion activator 2
chr14_-_77028663
Show fit
1.00
ENST00000238647.5
IRF2BPL
interferon regulatory factor 2 binding protein like
chr1_-_109619605
Show fit
0.98
ENST00000679935.1
GNAT2
G protein subunit alpha transducin 2
chr6_+_89562308
Show fit
0.92
ENST00000522441.5
ANKRD6
ankyrin repeat domain 6
chr11_+_44726811
Show fit
0.92
ENST00000533202.5
ENST00000520358.7
ENST00000533080.5
ENST00000520999.6
TSPAN18
tetraspanin 18
chr1_+_222618075
Show fit
0.92
ENST00000344922.10
MIA3
MIA SH3 domain ER export factor 3
chr6_+_135851681
Show fit
0.91
ENST00000308191.11
PDE7B
phosphodiesterase 7B
chr4_-_10116324
Show fit
0.90
ENST00000508079.1
WDR1
WD repeat domain 1
chr5_+_74715503
Show fit
0.88
ENST00000513336.5
HEXB
hexosaminidase subunit beta
chr16_-_29745951
Show fit
0.87
ENST00000329410.4
C16orf54
chromosome 16 open reading frame 54
chr10_+_95755737
Show fit
0.86
ENST00000543964.6
ENTPD1
ectonucleoside triphosphate diphosphohydrolase 1
chr6_-_27472681
Show fit
0.86
ENST00000377419.1
ZNF184
zinc finger protein 184
chr17_+_65137408
Show fit
0.82
ENST00000443584.7
ENST00000449996.7
RGS9
regulator of G protein signaling 9
chr1_-_154956086
Show fit
0.81
ENST00000368463.8
ENST00000368460.7
ENST00000368465.5
PBXIP1
PBX homeobox interacting protein 1
chr10_+_95755652
Show fit
0.80
ENST00000371207.8
ENTPD1
ectonucleoside triphosphate diphosphohydrolase 1
chr15_-_29822028
Show fit
0.80
ENST00000545208.6
TJP1
tight junction protein 1
chr4_+_86934976
Show fit
0.80
ENST00000507468.5
ENST00000395146.9
AFF1
AF4/FMR2 family member 1
chr6_-_31728877
Show fit
0.79
ENST00000437288.5
DDAH2
dimethylarginine dimethylaminohydrolase 2
chr17_+_65137344
Show fit
0.79
ENST00000262406.10
RGS9
regulator of G protein signaling 9
chr16_+_67029359
Show fit
0.79
ENST00000565389.1
CBFB
core-binding factor subunit beta
chr4_+_119212636
Show fit
0.76
ENST00000274030.10
USP53
ubiquitin specific peptidase 53
chr6_-_31729785
Show fit
0.76
ENST00000416410.6
DDAH2
dimethylarginine dimethylaminohydrolase 2
chr6_-_77463485
Show fit
0.75
ENST00000369947.5
HTR1B
5-hydroxytryptamine receptor 1B
chr6_-_31729478
Show fit
0.73
ENST00000436437.2
DDAH2
dimethylarginine dimethylaminohydrolase 2
chr17_+_80220406
Show fit
0.71
ENST00000573809.5
ENST00000361193.8
ENST00000574967.5
ENST00000576126.5
ENST00000411502.7
ENST00000546047.6
ENST00000572725.5
SLC26A11
solute carrier family 26 member 11
chr6_-_31729260
Show fit
0.70
ENST00000375789.7
DDAH2
dimethylarginine dimethylaminohydrolase 2
chr19_-_42255119
Show fit
0.70
ENST00000222329.9
ENST00000594664.1
ERF
ENSG00000268643.2
ETS2 repressor factor
chr1_-_3611470
Show fit
0.70
ENST00000356575.9
MEGF6
multiple EGF like domains 6
chr8_-_90645512
Show fit
0.69
ENST00000422900.1
TMEM64
transmembrane protein 64
chr20_-_23421409
Show fit
0.68
ENST00000377026.4
ENST00000398425.7
ENST00000432543.6
ENST00000617876.4
NAPB
NSF attachment protein beta
chr6_-_27473058
Show fit
0.68
ENST00000683788.1
ENST00000211936.10
ZNF184
zinc finger protein 184
chr11_+_842807
Show fit
0.68
ENST00000397411.6
TSPAN4
tetraspanin 4
chr11_+_61816249
Show fit
0.68
ENST00000257261.10
FADS2
fatty acid desaturase 2
chr8_-_132760548
Show fit
0.67
ENST00000519187.5
ENST00000523829.5
ENST00000677595.1
ENST00000356838.7
ENST00000377901.8
ENST00000519304.1
TMEM71
transmembrane protein 71
chr15_-_34336749
Show fit
0.66
ENST00000397707.6
ENST00000560611.5
SLC12A6
solute carrier family 12 member 6
chr14_+_99793375
Show fit
0.66
ENST00000262233.11
ENST00000556714.5
EML1
EMAP like 1
chr18_-_55587335
Show fit
0.65
ENST00000638154.3
TCF4
transcription factor 4
chr2_-_19990058
Show fit
0.65
ENST00000281405.9
ENST00000345530.8
WDR35
WD repeat domain 35
chr1_-_9069797
Show fit
0.65
ENST00000473209.1
SLC2A5
solute carrier family 2 member 5
chr7_+_130486324
Show fit
0.63
ENST00000427521.6
ENST00000378576.9
MEST
mesoderm specific transcript
chr19_+_13906255
Show fit
0.62
ENST00000589606.5
CC2D1A
coiled-coil and C2 domain containing 1A
chrX_-_31266857
Show fit
0.61
ENST00000378702.8
ENST00000361471.8
DMD
dystrophin
chr7_+_130486171
Show fit
0.60
ENST00000341441.9
ENST00000416162.7
MEST
mesoderm specific transcript
chr15_-_34337462
Show fit
0.59
ENST00000676379.1
SLC12A6
solute carrier family 12 member 6
chr12_-_53677397
Show fit
0.58
ENST00000338662.6
ENST00000552242.5
ATP5MC2
ATP synthase membrane subunit c locus 2
chr9_+_32551670
Show fit
0.57
ENST00000450093.3
SMIM27
small integral membrane protein 27
chr3_+_141426108
Show fit
0.57
ENST00000441582.2
ENST00000510726.1
ZBTB38
zinc finger and BTB domain containing 38
chr1_+_179882040
Show fit
0.56
ENST00000528443.6
TOR1AIP1
torsin 1A interacting protein 1
chr10_+_75431605
Show fit
0.55
ENST00000611255.5
LRMDA
leucine rich melanocyte differentiation associated
chr12_-_13000166
Show fit
0.55
ENST00000647702.1
HEBP1
heme binding protein 1
chr5_-_147831627
Show fit
0.55
ENST00000510027.2
SPINK1
serine peptidase inhibitor Kazal type 1
chr22_+_44180915
Show fit
0.53
ENST00000444313.8
ENST00000416291.5
PARVG
parvin gamma
chr5_-_147831663
Show fit
0.53
ENST00000296695.10
SPINK1
serine peptidase inhibitor Kazal type 1
chrY_-_6911752
Show fit
0.52
ENST00000651267.2
AMELY
amelogenin Y-linked
chr10_-_100185993
Show fit
0.52
ENST00000421367.7
ENST00000370408.2
ENST00000407654.7
ERLIN1
ER lipid raft associated 1
chr3_-_72446623
Show fit
0.51
ENST00000477973.4
RYBP
RING1 and YY1 binding protein
chr7_-_122886439
Show fit
0.51
ENST00000412584.6
ENST00000449022.7
CADPS2
calcium dependent secretion activator 2
chr12_-_13000208
Show fit
0.50
ENST00000014930.9
ENST00000536942.1
HEBP1
heme binding protein 1
chr13_+_48976597
Show fit
0.50
ENST00000541916.5
FNDC3A
fibronectin type III domain containing 3A
chr17_+_79730937
Show fit
0.50
ENST00000328313.10
ENPP7
ectonucleotide pyrophosphatase/phosphodiesterase 7
chr2_+_69829630
Show fit
0.50
ENST00000282570.4
GMCL1
germ cell-less 1, spermatogenesis associated
chr3_+_49007365
Show fit
0.49
ENST00000608424.6
ENST00000438660.5
ENST00000415265.6
WDR6
WD repeat domain 6
chr11_+_842824
Show fit
0.49
ENST00000397396.5
ENST00000397397.7
TSPAN4
tetraspanin 4
chr15_-_82349437
Show fit
0.49
ENST00000621197.4
ENST00000610657.2
ENST00000619556.4
GOLGA6L10
golgin A6 family like 10
chr1_+_179882275
Show fit
0.48
ENST00000606911.7
ENST00000271583.7
TOR1AIP1
torsin 1A interacting protein 1
chr12_-_70754631
Show fit
0.48
ENST00000440835.6
ENST00000549308.5
ENST00000550661.1
ENST00000378778.5
PTPRR
protein tyrosine phosphatase receptor type R
chr5_-_115296610
Show fit
0.48
ENST00000379611.10
ENST00000506442.5
CCDC112
coiled-coil domain containing 112
chr5_-_151686908
Show fit
0.47
ENST00000231061.9
SPARC
secreted protein acidic and cysteine rich
chr13_+_31739520
Show fit
0.47
ENST00000298386.7
RXFP2
relaxin family peptide receptor 2
chrX_-_31266925
Show fit
0.47
ENST00000680557.1
ENST00000378680.6
ENST00000378723.7
ENST00000680768.2
ENST00000681870.1
ENST00000680355.1
ENST00000682322.1
ENST00000682600.1
ENST00000680162.2
ENST00000681153.1
ENST00000681334.1
DMD
dystrophin
chr13_+_31739542
Show fit
0.46
ENST00000380314.2
RXFP2
relaxin family peptide receptor 2
chr11_+_842928
Show fit
0.46
ENST00000397408.5
TSPAN4
tetraspanin 4
chr11_-_18726187
Show fit
0.45
ENST00000513874.6
IGSF22
immunoglobulin superfamily member 22
chr16_-_50368920
Show fit
0.45
ENST00000394688.8
BRD7
bromodomain containing 7
chr4_+_86935040
Show fit
0.45
ENST00000674009.1
ENST00000503477.5
AFF1
AF4/FMR2 family member 1
chr4_-_139302516
Show fit
0.43
ENST00000394228.5
ENST00000539387.5
NDUFC1
NADH:ubiquinone oxidoreductase subunit C1
chr7_-_127392687
Show fit
0.43
ENST00000393313.5
ENST00000619291.4
ENST00000265827.8
ENST00000434602.5
ZNF800
zinc finger protein 800
chr19_+_40601342
Show fit
0.42
ENST00000396819.8
LTBP4
latent transforming growth factor beta binding protein 4
chr11_-_104164361
Show fit
0.42
ENST00000302251.9
PDGFD
platelet derived growth factor D
chr3_-_143848442
Show fit
0.41
ENST00000474151.1
ENST00000316549.11
SLC9A9
solute carrier family 9 member A9
chr3_+_49007062
Show fit
0.40
ENST00000395474.7
ENST00000610967.4
ENST00000429900.6
WDR6
WD repeat domain 6
chr15_+_84235773
Show fit
0.40
ENST00000510439.7
ENST00000422563.6
GOLGA6L4
golgin A6 family like 4
chr15_+_80059568
Show fit
0.40
ENST00000613266.4
ENST00000561060.5
ZFAND6
Links
zinc finger AN1-type containing 6
chr3_-_49813880
Show fit
0.39
ENST00000333486.4
UBA7
ubiquitin like modifier activating enzyme 7
chr8_+_81732434
Show fit
0.39
ENST00000297265.5
CHMP4C
charged multivesicular body protein 4C
chr16_-_50368779
Show fit
0.38
ENST00000394689.2
BRD7
bromodomain containing 7
chrX_+_51893533
Show fit
0.38
ENST00000375722.5
ENST00000326587.12
ENST00000375695.2
MAGED1
MAGE family member D1
chr11_-_27722021
Show fit
0.37
ENST00000314915.6
BDNF
brain derived neurotrophic factor
chr5_-_128537821
Show fit
0.36
ENST00000508989.5
FBN2
fibrillin 2
chr1_-_247172002
Show fit
0.36
ENST00000491356.5
ENST00000472531.5
ENST00000340684.10
ENST00000543802.3
ZNF124
zinc finger protein 124
chr15_+_76059973
Show fit
0.35
ENST00000388942.8
TMEM266
transmembrane protein 266
chr1_+_108746654
Show fit
0.34
ENST00000370008.4
STXBP3
syntaxin binding protein 3
chr1_-_25875669
Show fit
0.34
ENST00000675840.1
PAQR7
progestin and adipoQ receptor family member 7
chr4_+_6673940
Show fit
0.33
ENST00000635031.1
ENSG00000170846.17
novel protein, similar to Morf4 family associated protein 1
chr17_-_42136431
Show fit
0.33
ENST00000552162.5
ENST00000550504.5
RAB5C
RAB5C, member RAS oncogene family
chr6_+_28124596
Show fit
0.33
ENST00000340487.5
ZSCAN16
zinc finger and SCAN domain containing 16
chr2_-_61017174
Show fit
0.33
ENST00000407787.5
ENST00000398658.2
PUS10
pseudouridine synthase 10
chr8_-_90645905
Show fit
0.32
ENST00000418210.2
TMEM64
transmembrane protein 64
chr21_-_33479914
Show fit
0.32
ENST00000542230.7
TMEM50B
transmembrane protein 50B
chrX_+_71283186
Show fit
0.32
ENST00000535149.5
NONO
non-POU domain containing octamer binding
chr8_+_74320613
Show fit
0.31
ENST00000675821.1
GDAP1
ganglioside induced differentiation associated protein 1
chr15_-_29822077
Show fit
0.31
ENST00000677774.1
TJP1
tight junction protein 1
chr12_+_101594849
Show fit
0.31
ENST00000547405.5
ENST00000452455.6
ENST00000392934.7
ENST00000547509.5
ENST00000361685.6
ENST00000549145.5
ENST00000361466.7
ENST00000553190.5
ENST00000545503.6
ENST00000536007.5
ENST00000541119.5
ENST00000551300.5
ENST00000550270.1
MYBPC1
myosin binding protein C1
chr12_-_57430956
Show fit
0.31
ENST00000347140.7
ENST00000402412.5
R3HDM2
R3H domain containing 2
chr19_+_13906190
Show fit
0.31
ENST00000318003.11
CC2D1A
coiled-coil and C2 domain containing 1A
chr3_-_114624921
Show fit
0.30
ENST00000393785.6
ZBTB20
zinc finger and BTB domain containing 20
chr16_-_31428325
Show fit
0.30
ENST00000287490.5
COX6A2
cytochrome c oxidase subunit 6A2
chr20_+_31440626
Show fit
0.30
ENST00000376309.4
DEFB123
defensin beta 123
chr16_+_981762
Show fit
0.30
ENST00000293894.4
SOX8
SRY-box transcription factor 8
chr15_+_80059635
Show fit
0.29
ENST00000559157.5
ZFAND6
Links
zinc finger AN1-type containing 6
chr11_+_32091065
Show fit
0.28
ENST00000054950.4
RCN1
reticulocalbin 1
chr11_-_71448315
Show fit
0.28
ENST00000525346.5
ENST00000531364.5
ENST00000529990.5
ENST00000527316.5
ENST00000355527.8
ENST00000407721.6
DHCR7
7-dehydrocholesterol reductase
chr11_+_111976902
Show fit
0.27
ENST00000614104.4
DIXDC1
DIX domain containing 1
chr6_-_135497230
Show fit
0.27
ENST00000681365.1
AHI1
Abelson helper integration site 1
chr13_+_75788838
Show fit
0.26
ENST00000497947.6
LMO7
LIM domain 7
chr11_+_70270671
Show fit
0.23
ENST00000253925.12
ENST00000389547.7
PPFIA1
PTPRF interacting protein alpha 1
chr3_-_114624979
Show fit
0.23
ENST00000676079.1
ZBTB20
zinc finger and BTB domain containing 20
chr11_-_71448406
Show fit
0.23
ENST00000682708.1
ENST00000683287.1
ENST00000683714.1
ENST00000682880.1
DHCR7
7-dehydrocholesterol reductase
chr17_+_28744034
Show fit
0.23
ENST00000444415.7
ENST00000262396.10
TRAF4
TNF receptor associated factor 4
chr14_+_67619911
Show fit
0.23
ENST00000261783.4
ARG2
arginase 2
chr19_-_12513833
Show fit
0.23
ENST00000455490.1
ZNF709
zinc finger protein 709
chr1_+_43650118
Show fit
0.22
ENST00000372396.4
KDM4A
lysine demethylase 4A
chr12_-_122422544
Show fit
0.22
ENST00000358808.6
ENST00000361654.8
ENST00000539080.1
ENST00000537178.5
CLIP1
CAP-Gly domain containing linker protein 1
chr12_+_103587266
Show fit
0.22
ENST00000388887.7
STAB2
stabilin 2
chrX_-_100731504
Show fit
0.22
ENST00000372989.5
ENST00000276141.10
SYTL4
synaptotagmin like 4
chrX_+_52184874
Show fit
0.21
ENST00000599522.7
ENST00000471932.6
MAGED4
MAGE family member D4
chr15_-_44194407
Show fit
0.20
ENST00000484674.5
FRMD5
FERM domain containing 5
chr3_-_197298092
Show fit
0.20
ENST00000392382.6
DLG1
discs large MAGUK scaffold protein 1
chr12_-_21657792
Show fit
0.20
ENST00000396076.5
LDHB
lactate dehydrogenase B
chr12_-_21657831
Show fit
0.20
ENST00000450584.5
ENST00000350669.5
ENST00000673047.2
LDHB
lactate dehydrogenase B
chrX_+_52184904
Show fit
0.19
ENST00000375626.7
ENST00000467526.1
MAGED4
MAGE family member D4
chr5_+_138753412
Show fit
0.18
ENST00000520339.5
ENST00000627109.2
ENST00000302763.12
ENST00000518910.5
CTNNA1
catenin alpha 1
chr1_+_62437015
Show fit
0.18
ENST00000339950.5
USP1
ubiquitin specific peptidase 1
chr22_-_17258235
Show fit
0.18
ENST00000649310.1
ENST00000649746.1
ADA2
adenosine deaminase 2
chr22_-_23632317
Show fit
0.18
ENST00000317749.9
DRICH1
aspartate rich 1
chr6_-_56843638
Show fit
0.17
ENST00000421834.6
ENST00000370788.6
DST
dystonin
chr17_+_44141899
Show fit
0.17
ENST00000319977.8
ENST00000585683.6
HROB
homologous recombination factor with OB-fold
chr6_-_169723931
Show fit
0.17
ENST00000366780.8
ENST00000612128.1
PHF10
PHD finger protein 10
chr4_-_86934700
Show fit
0.17
ENST00000473559.5
ENSG00000285458.1
novel protein
chr14_+_105419938
Show fit
0.16
ENST00000405646.5
MTA1
metastasis associated 1
chrX_-_52069172
Show fit
0.16
ENST00000486010.1
ENST00000497164.5
ENST00000360134.10
ENST00000485287.5
ENST00000335504.9
MAGED4B
MAGE family member D4B
chr3_+_133746385
Show fit
0.16
ENST00000482271.5
ENST00000402696.9
TF
transferrin
chr17_-_29078857
Show fit
0.16
ENST00000359450.6
TIAF1
TGFB1-induced anti-apoptotic factor 1
chr1_+_103655760
Show fit
0.16
ENST00000370083.9
AMY1A
amylase alpha 1A
chr1_+_150549734
Show fit
0.16
ENST00000674043.1
ENST00000674058.1
ADAMTSL4
ADAMTS like 4
chr15_+_80060113
Show fit
0.15
ENST00000618205.4
ZFAND6
Links
zinc finger AN1-type containing 6
chr19_-_16627943
Show fit
0.15
ENST00000611692.4
MED26
mediator complex subunit 26
chr5_+_32710630
Show fit
0.15
ENST00000326958.5
NPR3
natriuretic peptide receptor 3
chr1_-_50960230
Show fit
0.15
ENST00000396153.7
FAF1
Fas associated factor 1
chr1_+_42682954
Show fit
0.15
ENST00000436427.1
YBX1
Y-box binding protein 1
chr8_-_33473076
Show fit
0.14
ENST00000524021.1
ENST00000327671.10
FUT10
fucosyltransferase 10
chr9_+_214843
Show fit
0.14
ENST00000432829.7
DOCK8
dedicator of cytokinesis 8
chr19_-_46471407
Show fit
0.14
ENST00000438932.2
PNMA8A
PNMA family member 8A
chr6_-_10419638
Show fit
0.13
ENST00000319516.8
TFAP2A
transcription factor AP-2 alpha
chr19_-_46471484
Show fit
0.13
ENST00000313683.15
ENST00000602246.1
PNMA8A
PNMA family member 8A
chr17_-_64263221
Show fit
0.13
ENST00000258991.7
ENST00000583738.1
ENST00000584379.6
TEX2
testis expressed 2
chr19_-_6459735
Show fit
0.13
ENST00000334510.9
ENST00000301454.9
SLC25A23
solute carrier family 25 member 23
chr4_-_140427635
Show fit
0.12
ENST00000325617.10
ENST00000414773.5
CLGN
calmegin
chr6_+_70413417
Show fit
0.12
ENST00000505769.5
ENST00000515323.5
FAM135A
family with sequence similarity 135 member A
chr11_+_7251886
Show fit
0.12
ENST00000318881.11
SYT9
synaptotagmin 9