Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
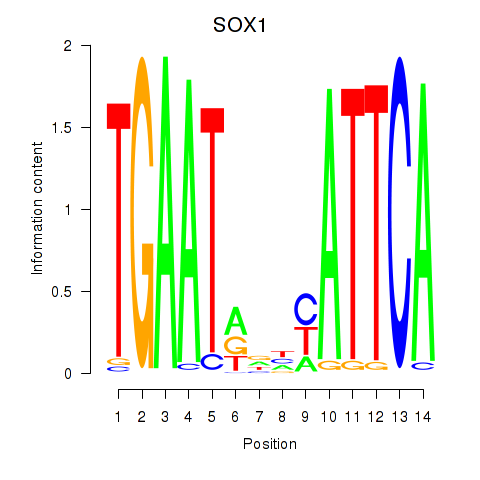
Results for SOX1
Z-value: 0.67
Transcription factors associated with SOX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX1
|
ENSG00000182968.5 | SOX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX1 | hg38_v1_chr13_+_112067143_112067160 | 0.00 | 9.9e-01 | Click! |
Activity profile of SOX1 motif
Sorted Z-values of SOX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_162318129 | 2.66 |
ENST00000679938.1
|
IFIH1
|
interferon induced with helicase C domain 1 |
| chr1_-_12616762 | 2.38 |
ENST00000464917.5
|
DHRS3
|
dehydrogenase/reductase 3 |
| chr19_-_10380558 | 1.99 |
ENST00000524462.5
ENST00000525621.6 ENST00000531836.5 |
TYK2
|
tyrosine kinase 2 |
| chr11_-_57410113 | 1.43 |
ENST00000529411.1
|
ENSG00000254979.5
|
novel protein |
| chr19_+_3762665 | 1.32 |
ENST00000330133.5
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr3_-_190122317 | 1.32 |
ENST00000427335.6
|
P3H2
|
prolyl 3-hydroxylase 2 |
| chr1_+_65147657 | 1.17 |
ENST00000546702.5
|
AK4
|
adenylate kinase 4 |
| chr2_-_162318475 | 1.16 |
ENST00000648433.1
|
IFIH1
|
interferon induced with helicase C domain 1 |
| chr5_-_16916400 | 1.10 |
ENST00000513882.5
|
MYO10
|
myosin X |
| chr7_-_106285898 | 1.08 |
ENST00000424768.2
ENST00000681255.1 |
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr18_+_24139053 | 1.06 |
ENST00000463087.5
ENST00000585037.5 ENST00000399496.8 ENST00000486759.6 ENST00000577705.1 ENST00000415309.6 ENST00000621648.4 ENST00000581397.5 |
CABYR
|
calcium binding tyrosine phosphorylation regulated |
| chr9_-_2844058 | 0.90 |
ENST00000397885.3
|
PUM3
|
pumilio RNA binding family member 3 |
| chr2_+_233388146 | 0.85 |
ENST00000409813.7
|
DGKD
|
diacylglycerol kinase delta |
| chr2_-_119366757 | 0.85 |
ENST00000414534.1
|
C2orf76
|
chromosome 2 open reading frame 76 |
| chr20_+_33217325 | 0.79 |
ENST00000375452.3
ENST00000375454.8 |
BPIFA3
|
BPI fold containing family A member 3 |
| chr10_-_90921079 | 0.77 |
ENST00000371697.4
|
ANKRD1
|
ankyrin repeat domain 1 |
| chr11_-_76670737 | 0.75 |
ENST00000260061.9
ENST00000404995.5 |
LRRC32
|
leucine rich repeat containing 32 |
| chr1_+_65147514 | 0.74 |
ENST00000545314.5
|
AK4
|
adenylate kinase 4 |
| chr4_+_155758990 | 0.69 |
ENST00000505154.5
ENST00000652626.1 ENST00000502959.5 ENST00000264424.13 ENST00000505764.5 ENST00000507146.5 ENST00000503520.5 |
GUCY1B1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr2_+_11724333 | 0.67 |
ENST00000425416.6
ENST00000396097.5 |
LPIN1
|
lipin 1 |
| chr19_+_3762705 | 0.66 |
ENST00000589174.1
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr3_-_16482850 | 0.66 |
ENST00000432519.5
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr9_-_83267230 | 0.64 |
ENST00000328788.5
|
FRMD3
|
FERM domain containing 3 |
| chr2_-_162318613 | 0.64 |
ENST00000649979.2
ENST00000421365.2 |
IFIH1
|
interferon induced with helicase C domain 1 |
| chr11_-_113706330 | 0.61 |
ENST00000540540.5
ENST00000538955.5 ENST00000545579.6 |
TMPRSS5
|
transmembrane serine protease 5 |
| chr10_-_43397220 | 0.58 |
ENST00000477108.5
ENST00000544000.5 |
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr11_-_113706292 | 0.57 |
ENST00000544634.5
ENST00000539732.5 ENST00000538770.1 ENST00000536856.5 ENST00000299882.11 ENST00000544476.1 |
TMPRSS5
|
transmembrane serine protease 5 |
| chr11_-_76669985 | 0.56 |
ENST00000407242.6
ENST00000421973.1 |
LRRC32
|
leucine rich repeat containing 32 |
| chr20_-_23605917 | 0.52 |
ENST00000376971.4
|
CST9
|
cystatin 9 |
| chr3_+_128051610 | 0.52 |
ENST00000464451.5
|
SEC61A1
|
SEC61 translocon subunit alpha 1 |
| chr14_-_24609660 | 0.51 |
ENST00000557220.6
ENST00000216338.9 ENST00000382548.4 |
GZMH
|
granzyme H |
| chr12_+_9827472 | 0.50 |
ENST00000617793.4
ENST00000617889.5 ENST00000354855.7 ENST00000279545.7 |
KLRF1
|
killer cell lectin like receptor F1 |
| chrX_+_11111291 | 0.50 |
ENST00000321143.8
ENST00000380762.5 ENST00000380763.7 |
HCCS
|
holocytochrome c synthase |
| chr11_-_133845495 | 0.50 |
ENST00000299140.8
ENST00000532889.1 |
SPATA19
|
spermatogenesis associated 19 |
| chr3_-_58627596 | 0.49 |
ENST00000474531.5
ENST00000465970.1 |
FAM107A
|
family with sequence similarity 107 member A |
| chr1_-_144456805 | 0.48 |
ENST00000581897.6
ENST00000577412.5 |
NBPF15
|
NBPF member 15 |
| chr10_-_72216267 | 0.47 |
ENST00000342444.8
ENST00000533958.1 ENST00000672957.1 ENST00000527593.5 ENST00000672940.1 ENST00000530461.5 ENST00000317168.11 ENST00000524829.5 |
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr13_+_108596152 | 0.46 |
ENST00000356711.7
ENST00000251041.10 |
MYO16
|
myosin XVI |
| chr1_-_183653307 | 0.45 |
ENST00000308641.6
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme catalytic polypeptide like 4 |
| chr20_-_14337602 | 0.45 |
ENST00000378053.3
ENST00000341420.5 |
FLRT3
|
fibronectin leucine rich transmembrane protein 3 |
| chr15_+_59611776 | 0.44 |
ENST00000396065.3
ENST00000560585.5 |
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr17_-_69150062 | 0.43 |
ENST00000522787.5
ENST00000521538.5 |
ABCA10
|
ATP binding cassette subfamily A member 10 |
| chr10_-_13972355 | 0.42 |
ENST00000264546.10
|
FRMD4A
|
FERM domain containing 4A |
| chr4_-_170003738 | 0.42 |
ENST00000502832.1
ENST00000393704.3 |
MFAP3L
|
microfibril associated protein 3 like |
| chr1_-_200410001 | 0.42 |
ENST00000367353.2
|
ZNF281
|
zinc finger protein 281 |
| chr19_+_17719471 | 0.40 |
ENST00000600186.5
ENST00000597735.5 ENST00000324096.9 |
MAP1S
|
microtubule associated protein 1S |
| chr11_+_4704782 | 0.37 |
ENST00000380390.6
|
MMP26
|
matrix metallopeptidase 26 |
| chr20_+_50958805 | 0.37 |
ENST00000244051.3
|
MOCS3
|
molybdenum cofactor synthesis 3 |
| chr1_+_152985231 | 0.35 |
ENST00000368762.1
|
SPRR1A
|
small proline rich protein 1A |
| chr2_-_208190001 | 0.35 |
ENST00000451346.5
ENST00000341287.9 |
C2orf80
|
chromosome 2 open reading frame 80 |
| chr6_+_28281555 | 0.35 |
ENST00000259883.3
ENST00000682144.1 |
PGBD1
|
piggyBac transposable element derived 1 |
| chr10_-_72217126 | 0.35 |
ENST00000317126.8
|
ASCC1
|
activating signal cointegrator 1 complex subunit 1 |
| chr9_-_120876356 | 0.34 |
ENST00000456291.1
|
PHF19
|
PHD finger protein 19 |
| chr1_-_200410052 | 0.32 |
ENST00000294740.3
|
ZNF281
|
zinc finger protein 281 |
| chr12_+_92702843 | 0.28 |
ENST00000397833.3
|
PLEKHG7
|
pleckstrin homology and RhoGEF domain containing G7 |
| chr19_+_51761167 | 0.27 |
ENST00000340023.7
ENST00000599326.1 ENST00000598953.1 |
FPR2
|
formyl peptide receptor 2 |
| chrX_-_56995426 | 0.27 |
ENST00000640768.1
ENST00000638619.1 |
SPIN3
|
spindlin family member 3 |
| chr22_-_30266839 | 0.26 |
ENST00000403463.1
ENST00000215781.3 |
OSM
|
oncostatin M |
| chr11_+_62242232 | 0.26 |
ENST00000244926.4
|
SCGB1D2
|
secretoglobin family 1D member 2 |
| chr1_-_200409976 | 0.25 |
ENST00000367352.3
|
ZNF281
|
zinc finger protein 281 |
| chr1_-_52033772 | 0.25 |
ENST00000371614.2
|
KTI12
|
KTI12 chromatin associated homolog |
| chr11_-_26572130 | 0.24 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr6_-_130222813 | 0.23 |
ENST00000437477.6
ENST00000439090.7 |
SAMD3
|
sterile alpha motif domain containing 3 |
| chrX_-_56995508 | 0.23 |
ENST00000374919.6
ENST00000639007.1 ENST00000639583.1 ENST00000638289.1 ENST00000639525.1 ENST00000638386.1 |
SPIN3
|
spindlin family member 3 |
| chr14_-_73950393 | 0.23 |
ENST00000651776.1
|
FAM161B
|
FAM161 centrosomal protein B |
| chr3_+_178558700 | 0.22 |
ENST00000432997.5
ENST00000455865.5 |
KCNMB2
|
potassium calcium-activated channel subfamily M regulatory beta subunit 2 |
| chr1_-_43172504 | 0.22 |
ENST00000431635.6
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chr5_-_173616588 | 0.21 |
ENST00000285908.5
ENST00000311086.9 ENST00000480951.1 |
BOD1
|
biorientation of chromosomes in cell division 1 |
| chr17_-_31297231 | 0.20 |
ENST00000247271.5
|
OMG
|
oligodendrocyte myelin glycoprotein |
| chr17_-_4641670 | 0.20 |
ENST00000293761.8
|
ALOX15
|
arachidonate 15-lipoxygenase |
| chrX_+_129906146 | 0.20 |
ENST00000394422.8
|
UTP14A
|
UTP14A small subunit processome component |
| chr1_+_84479239 | 0.19 |
ENST00000370656.5
ENST00000370654.6 |
RPF1
|
ribosome production factor 1 homolog |
| chr6_-_70957029 | 0.19 |
ENST00000230053.11
|
B3GAT2
|
beta-1,3-glucuronyltransferase 2 |
| chr1_+_174799895 | 0.18 |
ENST00000489615.5
|
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr17_-_40665121 | 0.17 |
ENST00000394052.5
|
KRT222
|
keratin 222 |
| chr8_-_92017292 | 0.17 |
ENST00000521553.5
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1 |
| chr19_+_49085433 | 0.17 |
ENST00000221448.9
ENST00000598441.6 |
SNRNP70
|
small nuclear ribonucleoprotein U1 subunit 70 |
| chr3_+_138608575 | 0.16 |
ENST00000338446.8
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr1_+_240091866 | 0.16 |
ENST00000319653.14
|
FMN2
|
formin 2 |
| chr2_+_201116396 | 0.15 |
ENST00000395148.6
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chr1_+_203795614 | 0.15 |
ENST00000367210.3
ENST00000432282.5 ENST00000453771.5 ENST00000367214.5 ENST00000639812.1 ENST00000367212.7 ENST00000332127.8 ENST00000550078.2 |
ZC3H11A
ZBED6
|
zinc finger CCCH-type containing 11A zinc finger BED-type containing 6 |
| chr3_+_138608700 | 0.14 |
ENST00000360570.7
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr3_+_138608851 | 0.14 |
ENST00000393035.6
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr15_+_75903893 | 0.14 |
ENST00000565036.5
ENST00000569054.5 |
FBXO22
|
F-box protein 22 |
| chrX_+_129906118 | 0.13 |
ENST00000425117.6
|
UTP14A
|
UTP14A small subunit processome component |
| chr16_+_11965193 | 0.13 |
ENST00000053243.6
ENST00000396495.3 |
TNFRSF17
|
TNF receptor superfamily member 17 |
| chr10_+_76318330 | 0.13 |
ENST00000496424.2
|
LRMDA
|
leucine rich melanocyte differentiation associated |
| chr1_-_86383078 | 0.12 |
ENST00000460698.6
|
ODF2L
|
outer dense fiber of sperm tails 2 like |
| chr17_-_15265230 | 0.12 |
ENST00000676161.1
ENST00000646419.2 ENST00000312280.9 ENST00000494511.7 ENST00000580584.3 ENST00000676221.1 |
PMP22
|
peripheral myelin protein 22 |
| chr7_+_142749465 | 0.11 |
ENST00000486171.5
ENST00000619214.4 ENST00000311737.12 |
PRSS1
|
serine protease 1 |
| chr10_+_93993897 | 0.11 |
ENST00000371380.8
|
PLCE1
|
phospholipase C epsilon 1 |
| chr12_-_10454485 | 0.11 |
ENST00000408006.7
ENST00000544822.2 ENST00000536188.5 |
KLRC1
|
killer cell lectin like receptor C1 |
| chr11_-_26572102 | 0.10 |
ENST00000455601.6
|
MUC15
|
mucin 15, cell surface associated |
| chr10_-_95069489 | 0.10 |
ENST00000371270.6
ENST00000535898.5 ENST00000623108.3 |
CYP2C8
|
cytochrome P450 family 2 subfamily C member 8 |
| chrX_-_120878924 | 0.10 |
ENST00000613352.1
|
ENSG00000278646.1
|
novel protein similar to cancer/testis antigen family 47, member A12 (CT47A12) |
| chr1_+_12746192 | 0.10 |
ENST00000614859.5
|
C1orf158
|
chromosome 1 open reading frame 158 |
| chr20_-_50958520 | 0.09 |
ENST00000371588.10
ENST00000371584.9 ENST00000413082.1 ENST00000371582.8 |
DPM1
|
dolichyl-phosphate mannosyltransferase subunit 1, catalytic |
| chr3_-_185938006 | 0.08 |
ENST00000342294.4
ENST00000453386.7 ENST00000382191.4 |
TRA2B
|
transformer 2 beta homolog |
| chr6_+_26183750 | 0.08 |
ENST00000614097.3
|
H2BC6
|
H2B clustered histone 6 |
| chr2_+_101998955 | 0.07 |
ENST00000393414.6
|
IL1R2
|
interleukin 1 receptor type 2 |
| chr17_+_77185210 | 0.07 |
ENST00000431431.6
|
SEC14L1
|
SEC14 like lipid binding 1 |
| chr18_-_54224578 | 0.07 |
ENST00000583046.1
ENST00000398398.6 ENST00000256429.8 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr7_-_93219545 | 0.07 |
ENST00000440868.5
|
HEPACAM2
|
HEPACAM family member 2 |
| chr7_-_93219564 | 0.06 |
ENST00000341723.8
|
HEPACAM2
|
HEPACAM family member 2 |
| chr4_-_141212877 | 0.06 |
ENST00000420921.6
|
RNF150
|
ring finger protein 150 |
| chr20_-_45667177 | 0.06 |
ENST00000618797.4
|
WFDC11
|
WAP four-disulfide core domain 11 |
| chr11_-_47378391 | 0.06 |
ENST00000227163.8
|
SPI1
|
Spi-1 proto-oncogene |
| chr1_-_111563934 | 0.06 |
ENST00000443498.5
|
TMIGD3
|
transmembrane and immunoglobulin domain containing 3 |
| chr14_-_60165363 | 0.06 |
ENST00000557185.6
|
DHRS7
|
dehydrogenase/reductase 7 |
| chr7_-_43869491 | 0.05 |
ENST00000317534.6
ENST00000418740.5 |
MRPS24
|
mitochondrial ribosomal protein S24 |
| chr7_-_105389365 | 0.05 |
ENST00000482897.5
|
SRPK2
|
SRSF protein kinase 2 |
| chr15_+_40382715 | 0.05 |
ENST00000416151.6
ENST00000249776.12 |
KNSTRN
|
kinetochore localized astrin (SPAG5) binding protein |
| chr11_-_47378494 | 0.04 |
ENST00000533030.1
|
SPI1
|
Spi-1 proto-oncogene |
| chr20_-_50131125 | 0.04 |
ENST00000371656.3
|
PEDS1
|
plasmanylethanolamine desaturase 1 |
| chr11_+_125626229 | 0.04 |
ENST00000532449.6
ENST00000534070.5 |
CHEK1
|
checkpoint kinase 1 |
| chr15_-_45114149 | 0.04 |
ENST00000603300.1
ENST00000389039.11 |
DUOX2
|
dual oxidase 2 |
| chr17_-_17281232 | 0.04 |
ENST00000417352.5
ENST00000268717.10 |
COPS3
|
COP9 signalosome subunit 3 |
| chr3_+_11011640 | 0.04 |
ENST00000643396.1
|
SLC6A1
|
solute carrier family 6 member 1 |
| chr19_+_54874218 | 0.03 |
ENST00000355524.8
ENST00000391726.7 |
FCAR
|
Fc fragment of IgA receptor |
| chrX_-_72877864 | 0.02 |
ENST00000596389.5
|
DMRTC1
|
DMRT like family C1 |
| chr16_-_11782552 | 0.02 |
ENST00000396516.6
|
ZC3H7A
|
zinc finger CCCH-type containing 7A |
| chr2_+_170178136 | 0.02 |
ENST00000409044.7
ENST00000408978.9 |
MYO3B
|
myosin IIIB |
| chr4_-_73620629 | 0.02 |
ENST00000342081.7
|
RASSF6
|
Ras association domain family member 6 |
| chr14_-_20333268 | 0.02 |
ENST00000358932.9
ENST00000557665.5 |
CCNB1IP1
|
cyclin B1 interacting protein 1 |
| chr3_+_136957948 | 0.02 |
ENST00000329582.9
|
IL20RB
|
interleukin 20 receptor subunit beta |
| chr14_-_20333306 | 0.02 |
ENST00000353689.8
ENST00000437553.6 |
CCNB1IP1
|
cyclin B1 interacting protein 1 |
| chr17_-_81166160 | 0.02 |
ENST00000326724.9
|
AATK
|
apoptosis associated tyrosine kinase |
| chr3_+_174859315 | 0.01 |
ENST00000454872.6
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase like 2 |
| chr17_+_36064265 | 0.01 |
ENST00000616054.2
|
CCL18
|
C-C motif chemokine ligand 18 |
| chrX_-_116462965 | 0.00 |
ENST00000371894.5
|
CT83
|
cancer/testis antigen 83 |
| chr3_-_33645433 | 0.00 |
ENST00000635664.1
ENST00000485378.6 ENST00000313350.10 ENST00000487200.5 |
CLASP2
|
cytoplasmic linker associated protein 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.5 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.1 | 2.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 1.9 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.1 | 0.4 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.1 | 0.7 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 0.9 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.1 | 0.5 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.1 | 0.7 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.1 | 1.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.2 | GO:2001302 | regulation of engulfment of apoptotic cell(GO:1901074) lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.1 | 0.8 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 1.3 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 1.1 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.2 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.3 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.0 | 0.8 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 1.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.3 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.4 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 1.0 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.5 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 1.0 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.1 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.3 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.0 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.1 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 2.0 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 1.0 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 1.1 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.5 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.0 | 0.0 | GO:0051939 | gamma-aminobutyric acid import(GO:0051939) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.0 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0097229 | sperm end piece(GO:0097229) |
| 0.1 | 2.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 1.1 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 2.0 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.3 | 1.3 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.3 | 0.8 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 1.1 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.2 | 2.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.4 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.1 | 1.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.4 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 1.0 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 0.3 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.1 | 0.2 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.0 | 0.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.5 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 5.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.7 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.2 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 1.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.6 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.0 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 2.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.5 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 1.0 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |