Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
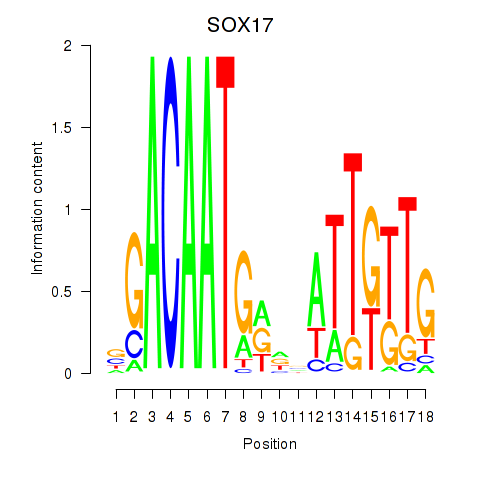
Results for SOX17
Z-value: 0.70
Transcription factors associated with SOX17
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX17
|
ENSG00000164736.6 | SOX17 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX17 | hg38_v1_chr8_+_54457927_54457943 | 0.22 | 2.8e-01 | Click! |
Activity profile of SOX17 motif
Sorted Z-values of SOX17 motif
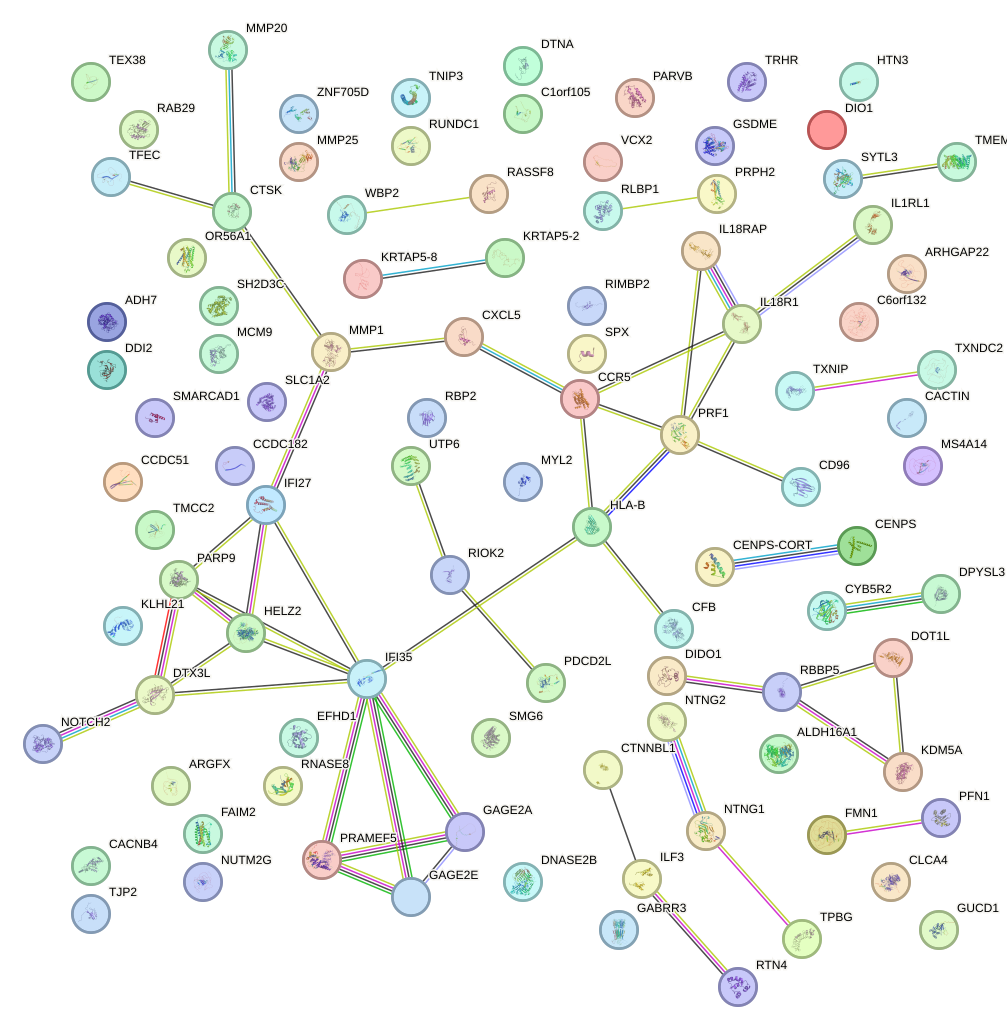
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX17
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_43006740 | 3.15 |
ENST00000438323.2
ENST00000415816.7 |
IFI35
|
interferon induced protein 35 |
| chr11_-_102798148 | 1.67 |
ENST00000315274.7
|
MMP1
|
matrix metallopeptidase 1 |
| chr4_-_73998669 | 1.44 |
ENST00000296027.5
|
CXCL5
|
C-X-C motif chemokine ligand 5 |
| chr20_-_63568074 | 1.38 |
ENST00000427522.6
|
HELZ2
|
helicase with zinc finger 2 |
| chr10_-_48605032 | 1.32 |
ENST00000249601.9
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr2_-_224947030 | 1.25 |
ENST00000409592.7
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr14_+_94110728 | 1.16 |
ENST00000616764.4
ENST00000618863.1 ENST00000611954.4 ENST00000618200.4 ENST00000621160.4 ENST00000555819.5 ENST00000620396.4 ENST00000612813.4 ENST00000620066.1 |
IFI27
|
interferon alpha inducible protein 27 |
| chr3_-_122564232 | 1.00 |
ENST00000471785.5
ENST00000466126.1 |
PARP9
|
poly(ADP-ribose) polymerase family member 9 |
| chr5_-_147453888 | 0.95 |
ENST00000398514.7
|
DPYSL3
|
dihydropyrimidinase like 3 |
| chr1_+_205256189 | 0.87 |
ENST00000329800.7
|
TMCC2
|
transmembrane and coiled-coil domain family 2 |
| chr17_+_42980547 | 0.86 |
ENST00000361677.5
ENST00000589705.1 |
RUNDC1
|
RUN domain containing 1 |
| chr1_+_86547070 | 0.85 |
ENST00000370563.3
|
CLCA4
|
chloride channel accessory 4 |
| chr11_-_7674206 | 0.82 |
ENST00000533558.5
ENST00000527542.5 |
CYB5R2
|
cytochrome b5 reductase 2 |
| chr7_-_115968302 | 0.81 |
ENST00000457268.5
|
TFEC
|
transcription factor EC |
| chr3_-_122564253 | 0.78 |
ENST00000492382.5
ENST00000682323.1 ENST00000462315.5 |
PARP9
|
poly(ADP-ribose) polymerase family member 9 |
| chr8_+_12104389 | 0.77 |
ENST00000400085.7
|
ZNF705D
|
zinc finger protein 705D |
| chr12_-_110920568 | 0.72 |
ENST00000548438.1
ENST00000228841.15 |
MYL2
|
myosin light chain 2 |
| chr18_+_34709356 | 0.71 |
ENST00000585446.1
ENST00000681241.1 |
DTNA
|
dystrobrevin alpha |
| chr3_+_122564327 | 0.70 |
ENST00000296161.9
ENST00000383661.3 |
DTX3L
|
deltex E3 ubiquitin ligase 3L |
| chrX_-_8171267 | 0.70 |
ENST00000317103.5
|
VCX2
|
variable charge X-linked 2 |
| chr9_+_96928310 | 0.67 |
ENST00000354649.7
|
NUTM2G
|
NUT family member 2G |
| chr1_+_172452885 | 0.66 |
ENST00000367725.4
|
C1orf105
|
chromosome 1 open reading frame 105 |
| chrX_+_49341192 | 0.65 |
ENST00000621907.1
|
GAGE2E
|
G antigen 2E |
| chr14_+_21057822 | 0.63 |
ENST00000308227.2
|
RNASE8
|
ribonuclease A family member 8 |
| chr2_+_102418642 | 0.63 |
ENST00000264260.6
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr2_+_102311502 | 0.57 |
ENST00000404917.6
ENST00000410040.5 |
IL1RL1
IL18R1
|
interleukin 1 receptor like 1 interleukin 18 receptor 1 |
| chr1_+_53894181 | 0.54 |
ENST00000361921.8
ENST00000322679.10 ENST00000613679.4 ENST00000617230.2 ENST00000610607.4 ENST00000532493.5 ENST00000525202.5 ENST00000524406.5 ENST00000388876.3 |
DIO1
|
iodothyronine deiodinase 1 |
| chr3_+_46370854 | 0.54 |
ENST00000292303.4
|
CCR5
|
C-C motif chemokine receptor 5 |
| chr11_-_6030758 | 0.53 |
ENST00000641900.1
|
OR56A1
|
olfactory receptor family 56 subfamily A member 1 |
| chr1_-_120054225 | 0.53 |
ENST00000602566.6
|
NOTCH2
|
notch receptor 2 |
| chr1_-_205121986 | 0.52 |
ENST00000367164.1
|
RBBP5
|
RB binding protein 5, histone lysine methyltransferase complex subunit |
| chr19_+_16661121 | 0.51 |
ENST00000187762.7
ENST00000599479.1 |
TMEM38A
|
transmembrane protein 38A |
| chr1_-_205121964 | 0.50 |
ENST00000264515.11
|
RBBP5
|
RB binding protein 5, histone lysine methyltransferase complex subunit |
| chr6_+_158649997 | 0.47 |
ENST00000360448.8
ENST00000367081.7 ENST00000611299.5 |
SYTL3
|
synaptotagmin like 3 |
| chr1_+_15617415 | 0.45 |
ENST00000480945.6
|
DDI2
|
DNA damage inducible 1 homolog 2 |
| chr8_+_109086585 | 0.45 |
ENST00000518632.2
|
TRHR
|
thyrotropin releasing hormone receptor |
| chr4_-_121164314 | 0.42 |
ENST00000057513.8
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr22_-_24555920 | 0.41 |
ENST00000621833.4
ENST00000404664.7 |
GUCD1
|
guanylyl cyclase domain containing 1 |
| chr9_-_127778659 | 0.41 |
ENST00000314830.13
|
SH2D3C
|
SH2 domain containing 3C |
| chr9_+_69123009 | 0.41 |
ENST00000647986.1
|
TJP2
|
tight junction protein 2 |
| chr16_+_33827140 | 0.40 |
ENST00000562905.2
|
IGHV3OR16-13
|
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
| chr6_-_31357171 | 0.40 |
ENST00000412585.7
ENST00000434333.1 |
HLA-B
|
major histocompatibility complex, class I, B |
| chr15_-_89211803 | 0.39 |
ENST00000563254.1
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr22_-_24555086 | 0.38 |
ENST00000447813.6
ENST00000402766.5 ENST00000435822.6 ENST00000407471.7 |
GUCD1
|
guanylyl cyclase domain containing 1 |
| chr4_+_70028452 | 0.38 |
ENST00000530128.5
ENST00000381057.3 ENST00000673563.1 |
HTN3
|
histatin 3 |
| chr11_-_35418966 | 0.38 |
ENST00000531628.2
|
SLC1A2
|
solute carrier family 1 member 2 |
| chr6_+_31948956 | 0.37 |
ENST00000483004.1
|
CFB
|
complement factor B |
| chr1_+_13254212 | 0.37 |
ENST00000622421.2
|
PRAMEF5
|
PRAME family member 5 |
| chr17_-_57745292 | 0.37 |
ENST00000299415.3
|
CCDC182
|
coiled-coil domain containing 182 |
| chr17_-_75855204 | 0.36 |
ENST00000589642.5
ENST00000593002.1 ENST00000590221.5 ENST00000587374.5 ENST00000585462.5 ENST00000254806.8 ENST00000433525.6 ENST00000626827.2 |
WBP2
|
WW domain binding protein 2 |
| chrY_-_23694579 | 0.36 |
ENST00000343584.10
|
PRYP3
|
PTPN13 like Y-linked pseudogene 3 |
| chr3_+_111542178 | 0.36 |
ENST00000283285.10
ENST00000352690.9 |
CD96
|
CD96 molecule |
| chr15_-_33067884 | 0.35 |
ENST00000334528.13
|
FMN1
|
formin 1 |
| chr19_+_32345593 | 0.35 |
ENST00000311921.8
ENST00000355898.6 ENST00000544431.5 |
ZNF507
|
zinc finger protein 507 |
| chr1_-_6602885 | 0.35 |
ENST00000377663.3
|
KLHL21
|
kelch like family member 21 |
| chr1_+_107139996 | 0.34 |
ENST00000370073.6
ENST00000370074.8 |
NTNG1
|
netrin G1 |
| chr1_+_84408230 | 0.34 |
ENST00000370662.3
|
DNASE2B
|
deoxyribonuclease 2 beta |
| chr6_+_82363199 | 0.34 |
ENST00000535040.4
|
TPBG
|
trophoblast glycoprotein |
| chr14_-_106557465 | 0.33 |
ENST00000390625.3
|
IGHV3-49
|
immunoglobulin heavy variable 3-49 |
| chr19_+_10654261 | 0.32 |
ENST00000449870.5
|
ILF3
|
interleukin enhancer binding factor 3 |
| chr1_+_10430070 | 0.32 |
ENST00000400900.6
|
CENPS-CORT
|
CENPS-CORT readthrough |
| chr1_-_150808251 | 0.31 |
ENST00000271651.8
ENST00000676970.1 ENST00000679260.1 ENST00000676751.1 ENST00000677887.1 |
CTSK
|
cathepsin K |
| chr2_+_232662733 | 0.31 |
ENST00000410095.5
ENST00000611312.1 |
EFHD1
|
EF-hand domain family member D1 |
| chr3_-_48440022 | 0.31 |
ENST00000447018.5
ENST00000442740.1 ENST00000412398.6 ENST00000395694.7 |
CCDC51
|
coiled-coil domain containing 51 |
| chr5_-_97183203 | 0.31 |
ENST00000508447.1
ENST00000283109.8 |
RIOK2
|
RIO kinase 2 |
| chr1_-_205775182 | 0.31 |
ENST00000446390.6
|
RAB29
|
RAB29, member RAS oncogene family |
| chr12_-_49897056 | 0.30 |
ENST00000552863.5
|
FAIM2
|
Fas apoptotic inhibitory molecule 2 |
| chr17_-_4949037 | 0.30 |
ENST00000572383.1
|
PFN1
|
profilin 1 |
| chr5_+_168486462 | 0.30 |
ENST00000231572.8
ENST00000626454.1 |
RARS1
|
arginyl-tRNA synthetase 1 |
| chr18_+_34710249 | 0.29 |
ENST00000680346.1
ENST00000348997.9 ENST00000681274.1 ENST00000680822.1 ENST00000680767.2 ENST00000597599.5 ENST00000444659.6 |
DTNA
|
dystrobrevin alpha |
| chr2_-_70995336 | 0.29 |
ENST00000606025.5
|
ENSG00000258881.6
|
novel protein |
| chr3_-_98035295 | 0.28 |
ENST00000621172.4
|
GABRR3
|
gamma-aminobutyric acid type A receptor subunit rho3 |
| chr1_+_46671871 | 0.28 |
ENST00000564373.1
|
TEX38
|
testis expressed 38 |
| chr7_-_24757926 | 0.28 |
ENST00000342947.9
ENST00000419307.6 |
GSDME
|
gasdermin E |
| chr11_-_102625332 | 0.28 |
ENST00000260228.3
|
MMP20
|
matrix metallopeptidase 20 |
| chr11_+_60396435 | 0.28 |
ENST00000395005.6
|
MS4A14
|
membrane spanning 4-domains A14 |
| chr17_-_31901658 | 0.28 |
ENST00000261708.9
|
UTP6
|
UTP6 small subunit processome component |
| chr3_+_121567924 | 0.27 |
ENST00000334384.5
|
ARGFX
|
arginine-fifty homeobox |
| chr19_+_2163915 | 0.27 |
ENST00000398665.8
|
DOT1L
|
DOT1 like histone lysine methyltransferase |
| chr22_+_44069043 | 0.27 |
ENST00000404989.1
|
PARVB
|
parvin beta |
| chr17_-_2266131 | 0.27 |
ENST00000570606.5
ENST00000354901.8 |
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr4_-_99144238 | 0.26 |
ENST00000512499.5
ENST00000504125.1 ENST00000505590.5 ENST00000629236.2 ENST00000508393.5 ENST00000265512.12 |
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr4_+_94207845 | 0.26 |
ENST00000457823.6
ENST00000354268.9 |
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr6_-_42142604 | 0.26 |
ENST00000356542.5
ENST00000341865.9 |
C6orf132
|
chromosome 6 open reading frame 132 |
| chr12_+_26011713 | 0.26 |
ENST00000542004.5
|
RASSF8
|
Ras association domain family member 8 |
| chr6_-_118935146 | 0.26 |
ENST00000619706.5
ENST00000316316.10 |
MCM9
|
minichromosome maintenance 9 homologous recombination repair factor |
| chr19_+_34404379 | 0.26 |
ENST00000246535.4
|
PDCD2L
|
programmed cell death 2 like |
| chr11_+_71538025 | 0.26 |
ENST00000398534.4
|
KRTAP5-8
|
keratin associated protein 5-8 |
| chrX_-_152709260 | 0.25 |
ENST00000535353.3
ENST00000638741.1 ENST00000640702.1 |
CSAG2
|
CSAG family member 2 |
| chr2_-_54987578 | 0.25 |
ENST00000486085.5
|
RTN4
|
reticulon 4 |
| chr12_+_21527017 | 0.25 |
ENST00000535033.5
|
SPX
|
spexin hormone |
| chr3_+_98168700 | 0.25 |
ENST00000383696.4
|
OR5H15
|
olfactory receptor family 5 subfamily H member 15 |
| chr6_-_42722590 | 0.25 |
ENST00000230381.7
|
PRPH2
|
peripherin 2 |
| chr20_+_37777262 | 0.25 |
ENST00000373469.1
|
CTNNBL1
|
catenin beta like 1 |
| chr19_+_49453211 | 0.25 |
ENST00000540132.5
ENST00000293350.9 ENST00000455361.6 |
ALDH16A1
|
aldehyde dehydrogenase 16 family member A1 |
| chr19_-_3626746 | 0.25 |
ENST00000429344.7
ENST00000248420.9 ENST00000221899.7 |
CACTIN
|
cactin, spliceosome C complex subunit |
| chr3_-_139476508 | 0.25 |
ENST00000232217.6
|
RBP2
|
retinol binding protein 2 |
| chr1_+_46671821 | 0.25 |
ENST00000334122.5
ENST00000415500.1 |
TEX38
|
testis expressed 38 |
| chr1_-_145995713 | 0.25 |
ENST00000425134.2
|
TXNIP
|
thioredoxin interacting protein |
| chr18_+_9885726 | 0.25 |
ENST00000611534.1
ENST00000536353.2 ENST00000584255.1 |
TXNDC2
|
thioredoxin domain containing 2 |
| chr2_-_151971750 | 0.24 |
ENST00000636598.1
|
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr18_+_58221535 | 0.24 |
ENST00000431212.6
ENST00000586268.5 ENST00000587190.5 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr17_-_3063607 | 0.24 |
ENST00000575751.1
|
OR1D5
|
olfactory receptor family 1 subfamily D member 5 |
| chr2_+_71130586 | 0.24 |
ENST00000498451.2
ENST00000244230.7 |
MPHOSPH10
|
M-phase phosphoprotein 10 |
| chr17_-_41489907 | 0.23 |
ENST00000328119.11
|
KRT36
|
keratin 36 |
| chr11_-_35419098 | 0.23 |
ENST00000606205.6
ENST00000645303.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr1_+_40374648 | 0.23 |
ENST00000372708.5
|
SMAP2
|
small ArfGAP2 |
| chr3_+_185586270 | 0.23 |
ENST00000296257.10
|
SENP2
|
SUMO specific peptidase 2 |
| chr1_+_47333863 | 0.23 |
ENST00000471289.2
ENST00000450808.2 |
CMPK1
|
cytidine/uridine monophosphate kinase 1 |
| chr6_+_29301701 | 0.23 |
ENST00000641895.1
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1 |
| chr1_-_6602859 | 0.23 |
ENST00000377658.8
|
KLHL21
|
kelch like family member 21 |
| chr6_-_118710065 | 0.22 |
ENST00000392500.7
ENST00000368488.9 ENST00000434604.5 |
CEP85L
|
centrosomal protein 85 like |
| chr22_-_17773976 | 0.22 |
ENST00000317361.11
|
BID
|
BH3 interacting domain death agonist |
| chr6_+_125956696 | 0.22 |
ENST00000229633.7
|
HINT3
|
histidine triad nucleotide binding protein 3 |
| chr1_+_47333774 | 0.22 |
ENST00000371873.10
|
CMPK1
|
cytidine/uridine monophosphate kinase 1 |
| chr2_-_89117844 | 0.22 |
ENST00000490686.1
|
IGKV1-17
|
immunoglobulin kappa variable 1-17 |
| chrX_-_13319952 | 0.22 |
ENST00000622204.1
ENST00000380622.5 |
ATXN3L
|
ataxin 3 like |
| chr6_-_52803807 | 0.22 |
ENST00000334575.6
|
GSTA1
|
glutathione S-transferase alpha 1 |
| chr1_-_13155961 | 0.22 |
ENST00000624207.1
|
PRAMEF26
|
PRAME family member 26 |
| chr6_+_135851681 | 0.22 |
ENST00000308191.11
|
PDE7B
|
phosphodiesterase 7B |
| chr6_+_125153649 | 0.22 |
ENST00000304877.17
ENST00000368402.9 ENST00000368388.6 ENST00000534000.6 |
TPD52L1
|
TPD52 like 1 |
| chr22_-_32255344 | 0.22 |
ENST00000266086.6
|
SLC5A4
|
solute carrier family 5 member 4 |
| chr8_+_109087424 | 0.21 |
ENST00000311762.2
|
TRHR
|
thyrotropin releasing hormone receptor |
| chr16_-_30012294 | 0.21 |
ENST00000564979.5
ENST00000563378.5 |
DOC2A
|
double C2 domain alpha |
| chr15_-_76012390 | 0.21 |
ENST00000394907.8
|
NRG4
|
neuregulin 4 |
| chr1_-_12945416 | 0.21 |
ENST00000415464.6
|
PRAMEF6
|
PRAME family member 6 |
| chr21_-_32603237 | 0.21 |
ENST00000431599.1
|
CFAP298
|
cilia and flagella associated protein 298 |
| chr1_-_120176450 | 0.20 |
ENST00000578049.4
|
SEC22B
|
SEC22 homolog B, vesicle trafficking protein |
| chr1_-_12886201 | 0.20 |
ENST00000235349.6
|
PRAMEF4
|
PRAME family member 4 |
| chr7_+_76424922 | 0.20 |
ENST00000394857.8
|
ZP3
|
zona pellucida glycoprotein 3 |
| chr19_-_49453472 | 0.20 |
ENST00000601825.1
ENST00000596049.5 ENST00000599366.5 ENST00000597415.5 |
PIH1D1
|
PIH1 domain containing 1 |
| chrX_+_8464830 | 0.20 |
ENST00000453306.4
ENST00000381032.6 ENST00000444481.3 |
VCX3B
|
variable charge X-linked 3B |
| chr6_+_31571957 | 0.20 |
ENST00000454783.5
|
LTA
|
lymphotoxin alpha |
| chr17_+_70104848 | 0.20 |
ENST00000392670.5
|
KCNJ16
|
potassium inwardly rectifying channel subfamily J member 16 |
| chrX_-_104013708 | 0.20 |
ENST00000217926.7
|
H2BW1
|
H2B.W histone 1 |
| chr11_+_5351508 | 0.20 |
ENST00000380219.1
|
OR51B6
|
olfactory receptor family 51 subfamily B member 6 |
| chr16_+_24846231 | 0.20 |
ENST00000568579.5
ENST00000567758.5 ENST00000569071.1 |
SLC5A11
|
solute carrier family 5 member 11 |
| chr7_+_135557904 | 0.19 |
ENST00000285968.11
|
NUP205
|
nucleoporin 205 |
| chr11_-_123885627 | 0.19 |
ENST00000528595.1
ENST00000375026.7 |
TMEM225
|
transmembrane protein 225 |
| chr1_+_13060769 | 0.19 |
ENST00000617807.3
|
HNRNPCL3
|
heterogeneous nuclear ribonucleoprotein C like 3 |
| chr2_+_86441341 | 0.19 |
ENST00000312912.10
ENST00000409064.5 |
KDM3A
|
lysine demethylase 3A |
| chrX_-_6535118 | 0.19 |
ENST00000381089.7
ENST00000612369.4 ENST00000398729.1 |
VCX3A
|
variable charge X-linked 3A |
| chr1_+_13315581 | 0.19 |
ENST00000376152.2
|
PRAMEF15
|
PRAME family member 15 |
| chr7_-_29146436 | 0.19 |
ENST00000396276.7
|
CPVL
|
carboxypeptidase vitellogenic like |
| chr9_-_136866265 | 0.19 |
ENST00000371648.4
ENST00000371649.5 |
EDF1
|
endothelial differentiation related factor 1 |
| chr6_-_125301900 | 0.18 |
ENST00000608295.5
ENST00000398153.7 ENST00000608284.1 |
HDDC2
|
HD domain containing 2 |
| chr19_+_41376499 | 0.18 |
ENST00000392002.7
|
TMEM91
|
transmembrane protein 91 |
| chr19_-_12919256 | 0.18 |
ENST00000293695.8
|
SYCE2
|
synaptonemal complex central element protein 2 |
| chr11_+_55885920 | 0.18 |
ENST00000244891.3
|
TRIM51
|
tripartite motif-containing 51 |
| chr1_+_14929734 | 0.18 |
ENST00000376028.8
ENST00000400798.6 |
KAZN
|
kazrin, periplakin interacting protein |
| chr17_-_75289212 | 0.18 |
ENST00000582778.1
ENST00000581988.5 ENST00000579207.5 ENST00000583332.5 ENST00000442286.6 ENST00000580151.5 ENST00000580994.5 ENST00000584438.1 ENST00000416858.7 ENST00000320362.7 |
SLC25A19
|
solute carrier family 25 member 19 |
| chr12_-_8662619 | 0.18 |
ENST00000544889.1
ENST00000543369.5 |
MFAP5
|
microfibril associated protein 5 |
| chr7_-_29146527 | 0.18 |
ENST00000265394.10
|
CPVL
|
carboxypeptidase vitellogenic like |
| chr8_-_33599935 | 0.18 |
ENST00000523956.1
ENST00000256261.9 |
DUSP26
|
dual specificity phosphatase 26 |
| chr8_+_117134989 | 0.17 |
ENST00000456015.7
|
SLC30A8
|
solute carrier family 30 member 8 |
| chr1_+_207496268 | 0.17 |
ENST00000529814.1
|
CR1
|
complement C3b/C4b receptor 1 (Knops blood group) |
| chr1_+_59310071 | 0.17 |
ENST00000371212.5
|
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr2_+_27628996 | 0.17 |
ENST00000616939.4
ENST00000264718.7 ENST00000610189.2 |
GPN1
|
GPN-loop GTPase 1 |
| chr11_+_60378524 | 0.17 |
ENST00000530614.5
ENST00000530027.5 ENST00000300184.8 ENST00000530234.2 ENST00000528215.1 ENST00000531787.5 |
MS4A7
MS4A14
|
membrane spanning 4-domains A7 membrane spanning 4-domains A14 |
| chr3_-_45796518 | 0.17 |
ENST00000413781.1
ENST00000358525.9 |
SLC6A20
|
solute carrier family 6 member 20 |
| chr12_-_76068933 | 0.17 |
ENST00000552056.5
|
NAP1L1
|
nucleosome assembly protein 1 like 1 |
| chr11_+_89924064 | 0.16 |
ENST00000623787.3
|
TRIM49D2
|
tripartite motif containing 49D2 |
| chr1_+_174875505 | 0.16 |
ENST00000486220.5
|
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr6_-_132659178 | 0.16 |
ENST00000275216.3
|
TAAR1
|
trace amine associated receptor 1 |
| chr19_-_53254841 | 0.16 |
ENST00000601828.5
ENST00000599012.5 ENST00000598513.6 ENST00000598806.5 |
ZNF677
|
zinc finger protein 677 |
| chr12_-_118359639 | 0.16 |
ENST00000541786.5
ENST00000419821.6 ENST00000541878.5 |
TAOK3
|
TAO kinase 3 |
| chr8_+_94641199 | 0.16 |
ENST00000646773.1
ENST00000454170.7 |
ESRP1
|
epithelial splicing regulatory protein 1 |
| chr11_+_60378475 | 0.16 |
ENST00000358246.5
|
MS4A7
|
membrane spanning 4-domains A7 |
| chr11_-_89922245 | 0.16 |
ENST00000420869.3
|
TRIM49D1
|
tripartite motif containing 49D1 |
| chr3_+_151733919 | 0.16 |
ENST00000356517.4
|
AADACL2
|
arylacetamide deacetylase like 2 |
| chr12_+_25052732 | 0.16 |
ENST00000547044.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr12_+_64824602 | 0.16 |
ENST00000539867.6
ENST00000544457.1 ENST00000539120.1 |
TBC1D30
|
TBC1 domain family member 30 |
| chr10_-_113854368 | 0.16 |
ENST00000369305.1
|
DCLRE1A
|
DNA cross-link repair 1A |
| chr7_+_99408609 | 0.16 |
ENST00000403633.6
|
BUD31
|
BUD31 homolog |
| chr7_-_122304738 | 0.16 |
ENST00000442488.7
|
FEZF1
|
FEZ family zinc finger 1 |
| chr10_-_49269 | 0.15 |
ENST00000562809.1
ENST00000568866.5 ENST00000561967.1 ENST00000568584.6 |
TUBB8
|
tubulin beta 8 class VIII |
| chr1_-_160523204 | 0.15 |
ENST00000368055.1
ENST00000368057.8 ENST00000368059.7 |
SLAMF6
|
SLAM family member 6 |
| chr12_-_11310420 | 0.15 |
ENST00000621732.4
ENST00000445719.2 ENST00000279575.7 |
PRB4
|
proline rich protein BstNI subfamily 4 |
| chr19_+_10654327 | 0.15 |
ENST00000407004.7
ENST00000589998.5 ENST00000589600.5 |
ILF3
|
interleukin enhancer binding factor 3 |
| chr1_+_13061158 | 0.15 |
ENST00000681473.1
|
HNRNPCL3
|
heterogeneous nuclear ribonucleoprotein C like 3 |
| chr1_-_13165631 | 0.15 |
ENST00000323770.8
|
HNRNPCL4
|
heterogeneous nuclear ribonucleoprotein C like 4 |
| chr7_+_148339452 | 0.15 |
ENST00000463592.3
|
CNTNAP2
|
contactin associated protein 2 |
| chr17_-_41168219 | 0.14 |
ENST00000391356.4
|
KRTAP4-3
|
keratin associated protein 4-3 |
| chr19_-_45406327 | 0.14 |
ENST00000593226.5
ENST00000418234.6 |
PPP1R13L
|
protein phosphatase 1 regulatory subunit 13 like |
| chr6_+_31572279 | 0.14 |
ENST00000418386.3
|
LTA
|
lymphotoxin alpha |
| chr3_+_178419123 | 0.14 |
ENST00000614557.1
ENST00000455307.5 ENST00000436432.1 |
ENSG00000275163.1
LINC01014
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 (KCNMB2-IT1 - KCNMB2 readthrough transcript) long intergenic non-protein coding RNA 1014 |
| chr7_+_95485934 | 0.14 |
ENST00000325885.6
|
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chrX_+_49331616 | 0.14 |
ENST00000612958.1
|
GAGE13
|
G antigen 13 |
| chr17_-_66229380 | 0.14 |
ENST00000205948.11
|
APOH
|
apolipoprotein H |
| chr4_-_149815826 | 0.14 |
ENST00000636793.2
ENST00000636414.1 |
IQCM
|
IQ motif containing M |
| chrX_+_120604084 | 0.14 |
ENST00000371317.10
|
MCTS1
|
MCTS1 re-initiation and release factor |
| chr9_-_128275935 | 0.14 |
ENST00000610329.4
|
GOLGA2
|
golgin A2 |
| chr2_-_100322495 | 0.14 |
ENST00000393437.8
|
LONRF2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr14_-_106088573 | 0.14 |
ENST00000632099.1
|
IGHV3-64D
|
immunoglobulin heavy variable 3-64D |
| chr1_+_13070853 | 0.13 |
ENST00000619661.2
|
PRAMEF25
|
PRAME family member 25 |
| chr12_-_11031407 | 0.13 |
ENST00000390675.2
|
TAS2R31
|
taste 2 receptor member 31 |
| chr1_+_171248471 | 0.13 |
ENST00000402921.6
ENST00000617670.6 ENST00000367750.7 |
FMO1
|
flavin containing dimethylaniline monoxygenase 1 |
| chr12_-_8662703 | 0.13 |
ENST00000535336.5
|
MFAP5
|
microfibril associated protein 5 |
| chr3_+_125969214 | 0.13 |
ENST00000508088.1
|
ROPN1B
|
rhophilin associated tail protein 1B |
| chr3_-_45796467 | 0.13 |
ENST00000353278.8
ENST00000456124.6 |
SLC6A20
|
solute carrier family 6 member 20 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.2 | 0.5 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.1 | 0.6 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.1 | 0.3 | GO:0002876 | positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 0.1 | 1.8 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.3 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.1 | 0.6 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) |
| 0.1 | 0.4 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 0.2 | GO:1903939 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 0.1 | 0.2 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.1 | 0.4 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 0.2 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 0.1 | 0.7 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.2 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 0.2 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 0.2 | GO:0045957 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) |
| 0.1 | 0.5 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.1 | 0.5 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.3 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.3 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.2 | GO:2000196 | positive regulation of female gonad development(GO:2000196) negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.5 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 1.4 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.6 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:1903238 | positive regulation of leukocyte tethering or rolling(GO:1903238) |
| 0.0 | 0.4 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.3 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 1.9 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 0.1 | GO:0071676 | negative regulation of mononuclear cell migration(GO:0071676) |
| 0.0 | 0.4 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.1 | GO:0002325 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.0 | 0.1 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.1 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.0 | 0.6 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.3 | GO:1904304 | regulation of gastro-intestinal system smooth muscle contraction(GO:1904304) positive regulation of gastro-intestinal system smooth muscle contraction(GO:1904306) |
| 0.0 | 4.6 | GO:0071357 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.2 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.0 | 0.1 | GO:0002769 | natural killer cell inhibitory signaling pathway(GO:0002769) |
| 0.0 | 0.1 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.2 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.2 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.0 | 0.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.3 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.0 | 0.3 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.1 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.0 | 0.1 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.0 | 0.1 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0032848 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.5 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.1 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.3 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.3 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.3 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.2 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.2 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 0.0 | 1.0 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.3 | GO:1903441 | protein localization to ciliary membrane(GO:1903441) |
| 0.0 | 0.3 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.2 | GO:0043697 | forebrain anterior/posterior pattern specification(GO:0021797) dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 1.1 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.6 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.0 | 0.1 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 0.0 | 0.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.3 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.9 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.1 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.6 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.1 | GO:0098704 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.2 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.8 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.1 | 0.7 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 1.0 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.3 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.1 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.3 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.1 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.3 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.2 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.2 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 1.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0042008 | interleukin-18 receptor activity(GO:0042008) |
| 0.1 | 0.3 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.1 | 0.5 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.6 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 1.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.5 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 1.0 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.4 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 0.8 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 1.8 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.2 | GO:0090422 | thiamine pyrophosphate transporter activity(GO:0090422) |
| 0.1 | 0.5 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.1 | 0.2 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.2 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.1 | 0.2 | GO:0004877 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.1 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.3 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 1.0 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.6 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.0 | 0.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 1.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.5 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.2 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.2 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.8 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.2 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.1 | GO:0032408 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.1 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0032190 | manganese ion transmembrane transporter activity(GO:0005384) acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 1.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.5 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.1 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 2.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.4 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 1.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 4.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 1.0 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |