Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for SOX30
Z-value: 0.53
Transcription factors associated with SOX30
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX30
|
ENSG00000039600.11 | SOX30 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX30 | hg38_v1_chr5_-_157652364_157652385, hg38_v1_chr5_-_157652432_157652443 | 0.05 | 8.2e-01 | Click! |
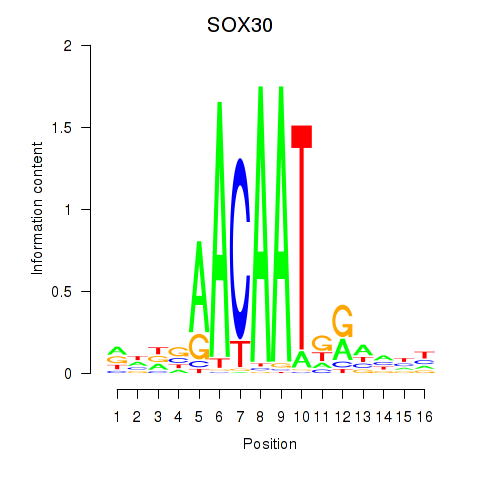
Activity profile of SOX30 motif
Sorted Z-values of SOX30 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX30
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_125919296 | 1.60 |
ENST00000444128.2
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr6_+_125919210 | 1.59 |
ENST00000438495.6
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr2_-_37672448 | 0.79 |
ENST00000611976.1
|
CDC42EP3
|
CDC42 effector protein 3 |
| chr2_-_37672178 | 0.74 |
ENST00000457889.1
|
CDC42EP3
|
CDC42 effector protein 3 |
| chr3_-_122564577 | 0.71 |
ENST00000477522.6
ENST00000360356.6 |
PARP9
|
poly(ADP-ribose) polymerase family member 9 |
| chr2_-_37671633 | 0.69 |
ENST00000295324.4
|
CDC42EP3
|
CDC42 effector protein 3 |
| chr5_-_147081428 | 0.68 |
ENST00000394413.7
|
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
| chr3_-_71306012 | 0.66 |
ENST00000649431.1
ENST00000610810.5 |
FOXP1
|
forkhead box P1 |
| chr2_+_27496830 | 0.63 |
ENST00000264717.7
|
GCKR
|
glucokinase regulator |
| chr7_+_101127095 | 0.59 |
ENST00000223095.5
|
SERPINE1
|
serpin family E member 1 |
| chr3_-_71305986 | 0.56 |
ENST00000647614.1
|
FOXP1
|
forkhead box P1 |
| chr11_-_85719160 | 0.54 |
ENST00000389958.7
ENST00000527794.5 |
SYTL2
|
synaptotagmin like 2 |
| chr11_-_85719045 | 0.51 |
ENST00000533057.6
ENST00000533892.5 |
SYTL2
|
synaptotagmin like 2 |
| chr17_+_4948252 | 0.48 |
ENST00000520221.5
|
ENO3
|
enolase 3 |
| chr5_-_147081462 | 0.46 |
ENST00000508267.5
ENST00000504198.5 |
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
| chr7_+_144000320 | 0.45 |
ENST00000641698.1
|
OR6B1
|
olfactory receptor family 6 subfamily B member 1 |
| chr11_-_85719111 | 0.45 |
ENST00000529581.5
ENST00000533577.1 |
SYTL2
|
synaptotagmin like 2 |
| chr11_-_35419462 | 0.42 |
ENST00000643522.1
|
SLC1A2
|
solute carrier family 1 member 2 |
| chr11_-_35420050 | 0.42 |
ENST00000395753.6
ENST00000395750.6 ENST00000645634.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr6_-_89217339 | 0.41 |
ENST00000454853.7
|
GABRR1
|
gamma-aminobutyric acid type A receptor subunit rho1 |
| chr11_-_35419213 | 0.40 |
ENST00000642171.1
ENST00000644050.1 ENST00000643134.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr6_-_139291987 | 0.40 |
ENST00000358430.8
|
TXLNB
|
taxilin beta |
| chr3_-_8644782 | 0.40 |
ENST00000544814.6
|
SSUH2
|
ssu-2 homolog |
| chr14_-_50561119 | 0.39 |
ENST00000555216.5
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr11_-_35420017 | 0.39 |
ENST00000643000.1
ENST00000646099.1 ENST00000647372.1 ENST00000642578.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr15_-_48963912 | 0.38 |
ENST00000332408.9
|
SHC4
|
SHC adaptor protein 4 |
| chr20_+_38962299 | 0.37 |
ENST00000373325.6
ENST00000373323.8 ENST00000252011.8 ENST00000615559.1 |
DHX35
ENSG00000277581.1
|
DEAH-box helicase 35 novel transcript, sense intronic to DHX35 |
| chr12_+_92702983 | 0.37 |
ENST00000344636.6
ENST00000544406.2 |
PLEKHG7
|
pleckstrin homology and RhoGEF domain containing G7 |
| chr12_+_92702843 | 0.36 |
ENST00000397833.3
|
PLEKHG7
|
pleckstrin homology and RhoGEF domain containing G7 |
| chr12_-_11395556 | 0.35 |
ENST00000565533.1
ENST00000389362.6 ENST00000546254.3 |
PRB2
PRB1
|
proline rich protein BstNI subfamily 2 proline rich protein BstNI subfamily 1 |
| chr11_+_54706832 | 0.35 |
ENST00000319760.8
|
OR4A5
|
olfactory receptor family 4 subfamily A member 5 |
| chr7_+_26152188 | 0.35 |
ENST00000056233.4
|
NFE2L3
|
nuclear factor, erythroid 2 like 3 |
| chr9_-_71768386 | 0.35 |
ENST00000377066.9
ENST00000377044.9 |
CEMIP2
|
cell migration inducing hyaluronidase 2 |
| chr11_+_128694052 | 0.34 |
ENST00000527786.7
ENST00000534087.3 |
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr11_+_128693887 | 0.31 |
ENST00000281428.12
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr8_-_81112055 | 0.30 |
ENST00000220597.4
|
PAG1
|
phosphoprotein membrane anchor with glycosphingolipid microdomains 1 |
| chr3_-_11643871 | 0.30 |
ENST00000430365.7
|
VGLL4
|
vestigial like family member 4 |
| chr16_+_3654683 | 0.29 |
ENST00000246949.10
|
DNASE1
|
deoxyribonuclease 1 |
| chr15_+_76336755 | 0.28 |
ENST00000290759.9
|
ISL2
|
ISL LIM homeobox 2 |
| chr5_+_141359970 | 0.27 |
ENST00000522605.2
ENST00000622527.1 |
PCDHGB2
|
protocadherin gamma subfamily B, 2 |
| chr5_+_141392616 | 0.26 |
ENST00000398604.3
|
PCDHGA8
|
protocadherin gamma subfamily A, 8 |
| chr12_-_49187369 | 0.26 |
ENST00000547939.6
|
TUBA1A
|
tubulin alpha 1a |
| chr11_-_35419899 | 0.25 |
ENST00000646847.1
ENST00000449068.2 ENST00000643401.1 ENST00000645966.1 ENST00000647104.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr1_-_167914089 | 0.24 |
ENST00000476818.2
ENST00000367848.1 ENST00000367851.9 ENST00000545172.5 |
ADCY10
|
adenylate cyclase 10 |
| chr6_+_131637296 | 0.24 |
ENST00000358229.6
ENST00000357639.8 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr3_-_167474026 | 0.23 |
ENST00000466903.1
ENST00000264677.8 |
SERPINI2
|
serpin family I member 2 |
| chr8_+_24294044 | 0.23 |
ENST00000265769.9
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr12_-_10098940 | 0.23 |
ENST00000420265.2
|
CLEC1A
|
C-type lectin domain family 1 member A |
| chr6_-_134318097 | 0.23 |
ENST00000367858.10
ENST00000533224.1 |
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr12_-_10098977 | 0.23 |
ENST00000315330.8
ENST00000457018.6 |
CLEC1A
|
C-type lectin domain family 1 member A |
| chr17_-_676348 | 0.23 |
ENST00000681510.1
ENST00000679680.1 |
VPS53
|
VPS53 subunit of GARP complex |
| chr5_-_160852200 | 0.22 |
ENST00000327245.10
|
ATP10B
|
ATPase phospholipid transporting 10B (putative) |
| chr1_-_201507116 | 0.22 |
ENST00000340006.7
ENST00000526256.5 ENST00000526723.5 ENST00000524951.5 |
CSRP1
|
cysteine and glycine rich protein 1 |
| chr4_-_175812746 | 0.22 |
ENST00000393658.6
|
GPM6A
|
glycoprotein M6A |
| chr2_-_165204042 | 0.22 |
ENST00000283254.12
ENST00000453007.1 |
SCN3A
|
sodium voltage-gated channel alpha subunit 3 |
| chr2_-_157325808 | 0.21 |
ENST00000410096.6
ENST00000420719.6 ENST00000409216.5 ENST00000419116.2 |
ERMN
|
ermin |
| chr12_-_98894830 | 0.21 |
ENST00000549797.5
ENST00000333732.11 ENST00000341752.11 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chrX_+_41333342 | 0.21 |
ENST00000629496.3
ENST00000625837.2 ENST00000626301.2 |
DDX3X
|
DEAD-box helicase 3 X-linked |
| chr16_+_4495852 | 0.21 |
ENST00000575129.5
ENST00000398595.7 ENST00000414777.5 |
HMOX2
|
heme oxygenase 2 |
| chr6_-_11778781 | 0.21 |
ENST00000414691.8
ENST00000229583.9 |
ADTRP
|
androgen dependent TFPI regulating protein |
| chr1_+_180928133 | 0.20 |
ENST00000367587.1
|
KIAA1614
|
KIAA1614 |
| chr11_+_196761 | 0.20 |
ENST00000325113.9
|
ODF3
|
outer dense fiber of sperm tails 3 |
| chr5_+_140855882 | 0.19 |
ENST00000562220.2
ENST00000307360.6 ENST00000506939.6 |
PCDHA10
|
protocadherin alpha 10 |
| chr2_-_165203870 | 0.18 |
ENST00000639244.1
ENST00000409101.7 ENST00000668657.1 |
SCN3A
|
sodium voltage-gated channel alpha subunit 3 |
| chr3_+_181711915 | 0.18 |
ENST00000325404.3
|
SOX2
|
SRY-box transcription factor 2 |
| chr7_-_93226449 | 0.18 |
ENST00000394468.7
ENST00000453812.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr6_-_127900958 | 0.17 |
ENST00000434358.3
ENST00000630369.2 ENST00000368248.4 |
THEMIS
|
thymocyte selection associated |
| chr14_+_51989609 | 0.17 |
ENST00000556760.5
|
RTRAF
|
RNA transcription, translation and transport factor |
| chr13_-_85799400 | 0.15 |
ENST00000647374.2
|
SLITRK6
|
SLIT and NTRK like family member 6 |
| chr17_-_75153826 | 0.15 |
ENST00000481647.5
ENST00000470924.5 |
JPT1
|
Jupiter microtubule associated homolog 1 |
| chr8_-_33567118 | 0.15 |
ENST00000256257.2
|
RNF122
|
ring finger protein 122 |
| chr7_-_83162899 | 0.15 |
ENST00000423517.6
|
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr17_-_40665121 | 0.14 |
ENST00000394052.5
|
KRT222
|
keratin 222 |
| chr7_+_144070313 | 0.14 |
ENST00000641441.1
|
OR2A25
|
olfactory receptor family 2 subfamily A member 25 |
| chr5_+_141408000 | 0.14 |
ENST00000616430.1
|
PCDHGB6
|
protocadherin gamma subfamily B, 6 |
| chr3_+_19947074 | 0.14 |
ENST00000273047.9
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr11_-_35419542 | 0.14 |
ENST00000643305.1
ENST00000644351.1 ENST00000278379.9 ENST00000644779.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr12_-_11310420 | 0.13 |
ENST00000621732.4
ENST00000445719.2 ENST00000279575.7 |
PRB4
|
proline rich protein BstNI subfamily 4 |
| chr5_+_141408032 | 0.12 |
ENST00000520790.1
|
PCDHGB6
|
protocadherin gamma subfamily B, 6 |
| chr14_+_51989508 | 0.12 |
ENST00000261700.8
|
RTRAF
|
RNA transcription, translation and transport factor |
| chr11_+_63369779 | 0.12 |
ENST00000279178.4
|
SLC22A9
|
solute carrier family 22 member 9 |
| chr12_-_95217373 | 0.12 |
ENST00000549499.1
ENST00000546711.5 ENST00000343958.9 |
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr9_+_100473140 | 0.11 |
ENST00000374879.5
|
TMEFF1
|
transmembrane protein with EGF like and two follistatin like domains 1 |
| chr2_+_222671651 | 0.11 |
ENST00000446656.4
|
MOGAT1
|
monoacylglycerol O-acyltransferase 1 |
| chr7_-_44189428 | 0.11 |
ENST00000673284.1
ENST00000403799.8 ENST00000671824.1 |
GCK
|
glucokinase |
| chr1_+_74235377 | 0.11 |
ENST00000326637.8
|
TNNI3K
|
TNNI3 interacting kinase |
| chrX_+_71283186 | 0.11 |
ENST00000535149.5
|
NONO
|
non-POU domain containing octamer binding |
| chr8_+_24294107 | 0.10 |
ENST00000437154.6
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr19_-_43935234 | 0.10 |
ENST00000269973.10
|
ZNF45
|
zinc finger protein 45 |
| chrX_+_71283577 | 0.10 |
ENST00000420903.6
ENST00000373856.8 ENST00000678437.1 ENST00000678830.1 ENST00000677879.1 ENST00000373841.5 ENST00000276079.13 ENST00000676797.1 ENST00000678660.1 ENST00000678231.1 ENST00000677612.1 ENST00000413858.5 ENST00000450092.6 |
NONO
|
non-POU domain containing octamer binding |
| chr7_+_142670734 | 0.09 |
ENST00000390398.3
|
TRBV25-1
|
T cell receptor beta variable 25-1 |
| chr22_+_50170720 | 0.09 |
ENST00000159647.9
ENST00000395842.3 |
PANX2
|
pannexin 2 |
| chrX_-_143636094 | 0.09 |
ENST00000356928.2
|
SLITRK4
|
SLIT and NTRK like family member 4 |
| chr5_+_66958870 | 0.08 |
ENST00000405643.5
ENST00000407621.1 ENST00000432426.5 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr9_-_83267230 | 0.08 |
ENST00000328788.5
|
FRMD3
|
FERM domain containing 3 |
| chr2_+_29113989 | 0.08 |
ENST00000404424.5
|
CLIP4
|
CAP-Gly domain containing linker protein family member 4 |
| chr8_+_104339796 | 0.08 |
ENST00000622554.1
ENST00000297581.2 |
DCSTAMP
|
dendrocyte expressed seven transmembrane protein |
| chr8_+_7836433 | 0.08 |
ENST00000314265.3
|
DEFB104A
|
defensin beta 104A |
| chr2_+_167135901 | 0.07 |
ENST00000628543.2
|
XIRP2
|
xin actin binding repeat containing 2 |
| chrX_+_70444827 | 0.07 |
ENST00000374360.8
|
DLG3
|
discs large MAGUK scaffold protein 3 |
| chr4_+_70592253 | 0.07 |
ENST00000322937.10
ENST00000613447.4 |
AMBN
|
ameloblastin |
| chr8_-_140800535 | 0.07 |
ENST00000521986.5
ENST00000523539.5 |
PTK2
|
protein tyrosine kinase 2 |
| chr2_+_99337364 | 0.07 |
ENST00000617677.1
ENST00000289371.11 |
EIF5B
|
eukaryotic translation initiation factor 5B |
| chr7_+_102285125 | 0.07 |
ENST00000536178.3
|
SH2B2
|
SH2B adaptor protein 2 |
| chrX_-_15854743 | 0.07 |
ENST00000450644.2
|
AP1S2
|
adaptor related protein complex 1 subunit sigma 2 |
| chr1_+_154429315 | 0.07 |
ENST00000476006.5
|
IL6R
|
interleukin 6 receptor |
| chr20_-_51768327 | 0.07 |
ENST00000311637.9
ENST00000338821.6 |
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr4_+_70592295 | 0.06 |
ENST00000449493.2
|
AMBN
|
ameloblastin |
| chr2_+_231708511 | 0.06 |
ENST00000341369.11
ENST00000409115.8 ENST00000409683.5 |
PTMA
|
prothymosin alpha |
| chrX_+_80420466 | 0.05 |
ENST00000308293.5
|
TENT5D
|
terminal nucleotidyltransferase 5D |
| chr12_-_68225806 | 0.05 |
ENST00000229134.5
|
IL26
|
interleukin 26 |
| chr12_-_11269805 | 0.05 |
ENST00000538488.2
|
PRB3
|
proline rich protein BstNI subfamily 3 |
| chr12_-_11269696 | 0.04 |
ENST00000381842.7
|
PRB3
|
proline rich protein BstNI subfamily 3 |
| chr16_-_2787550 | 0.04 |
ENST00000682474.1
|
PRSS33
|
serine protease 33 |
| chr1_-_118989504 | 0.03 |
ENST00000207157.7
|
TBX15
|
T-box transcription factor 15 |
| chr10_+_68721012 | 0.03 |
ENST00000536391.5
ENST00000630771.2 |
CCAR1
|
cell division cycle and apoptosis regulator 1 |
| chr1_+_240091866 | 0.03 |
ENST00000319653.14
|
FMN2
|
formin 2 |
| chr12_+_95217792 | 0.03 |
ENST00000436874.6
ENST00000551472.5 ENST00000552821.5 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr11_-_102705737 | 0.02 |
ENST00000260229.5
|
MMP27
|
matrix metallopeptidase 27 |
| chr1_+_244835616 | 0.02 |
ENST00000366528.3
ENST00000411948.7 |
COX20
|
cytochrome c oxidase assembly factor COX20 |
| chr1_+_185045515 | 0.01 |
ENST00000367509.8
ENST00000367510.8 |
RNF2
|
ring finger protein 2 |
| chr11_-_18526885 | 0.01 |
ENST00000251968.4
ENST00000536719.5 |
TSG101
|
tumor susceptibility 101 |
| chr12_+_10929229 | 0.01 |
ENST00000381847.7
ENST00000396400.4 |
PRH2
|
proline rich protein HaeIII subfamily 2 |
| chr7_-_149028452 | 0.01 |
ENST00000413966.1
ENST00000652332.1 |
PDIA4
|
protein disulfide isomerase family A member 4 |
| chr7_-_14903319 | 0.00 |
ENST00000403951.6
|
DGKB
|
diacylglycerol kinase beta |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0001300 | chronological cell aging(GO:0001300) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.2 | 3.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 2.0 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 2.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 1.5 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.3 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.2 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.6 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 1.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.7 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 1.1 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.1 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.6 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.2 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0034241 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.0 | 0.2 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 2.0 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 1.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.2 | GO:0001520 | outer dense fiber(GO:0001520) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.2 | 0.6 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 2.0 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 1.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.3 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 0.2 | GO:0043273 | CTPase activity(GO:0043273) |
| 0.0 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 3.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.7 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 1.2 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.7 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |