Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
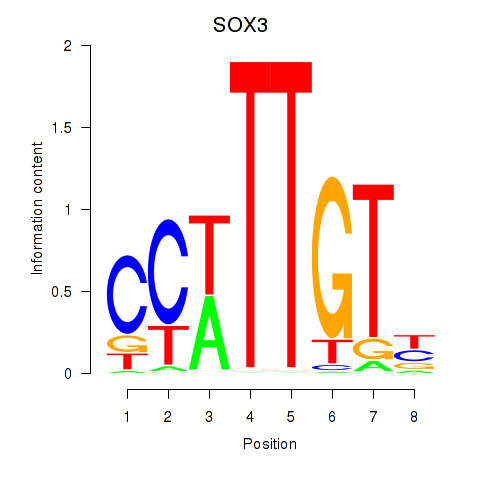
Results for SOX3_SOX2
Z-value: 2.13
Transcription factors associated with SOX3_SOX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX3
|
ENSG00000134595.9 | SOX3 |
|
SOX2
|
ENSG00000181449.4 | SOX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX3 | hg38_v1_chrX_-_140505058_140505076 | -0.35 | 8.6e-02 | Click! |
| SOX2 | hg38_v1_chr3_+_181711915_181711934 | 0.06 | 7.6e-01 | Click! |
Activity profile of SOX3_SOX2 motif
Sorted Z-values of SOX3_SOX2 motif
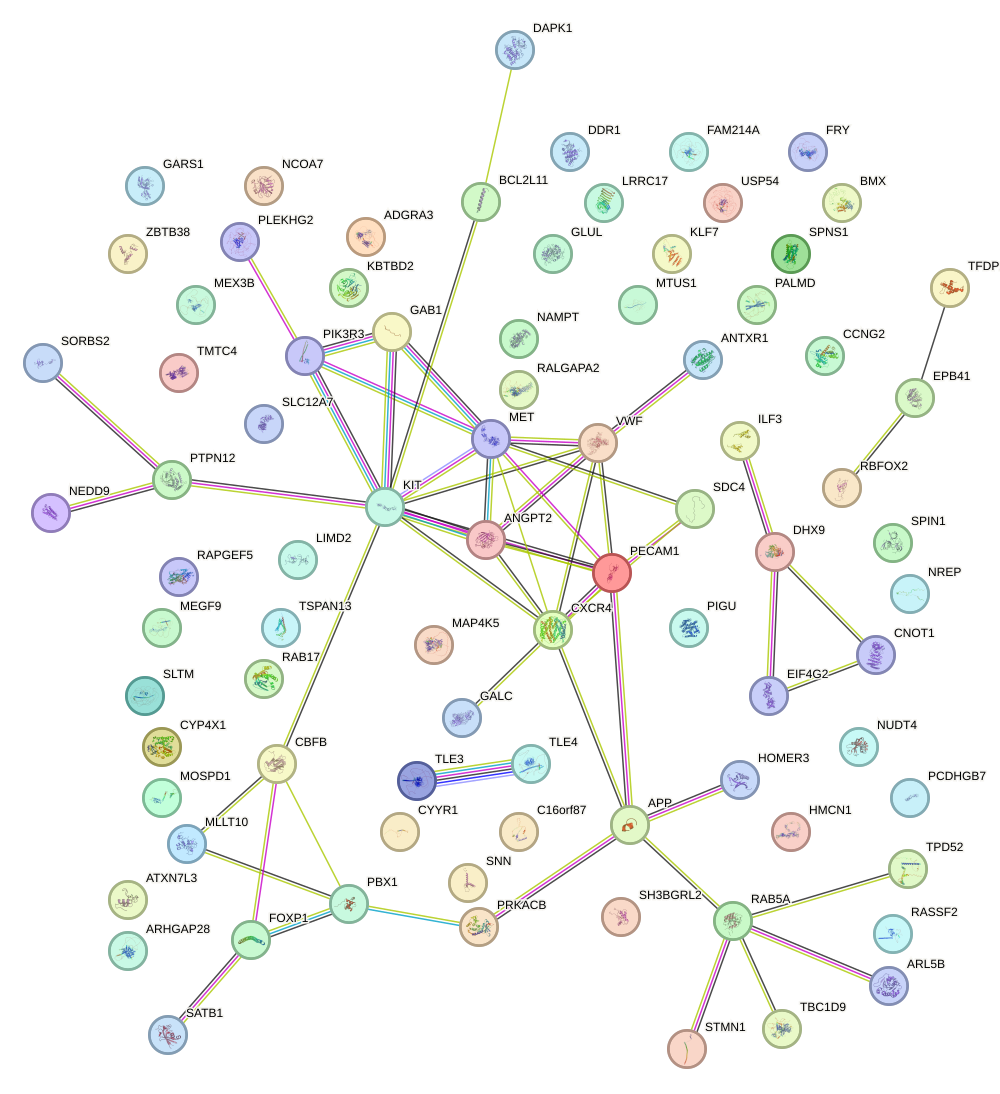
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX3_SOX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_10655023 | 8.27 |
ENST00000590009.5
|
ILF3
|
interleukin enhancer binding factor 3 |
| chr4_+_41612892 | 6.73 |
ENST00000509454.5
ENST00000396595.7 ENST00000381753.8 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr4_+_41612702 | 6.67 |
ENST00000509277.5
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr1_+_99646025 | 5.87 |
ENST00000263174.9
ENST00000605497.5 ENST00000615664.1 |
PALMD
|
palmdelphin |
| chr7_-_22193728 | 5.74 |
ENST00000620335.4
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr3_+_141386393 | 5.07 |
ENST00000503809.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr4_+_143381939 | 5.04 |
ENST00000505913.5
|
GAB1
|
GRB2 associated binding protein 1 |
| chr14_-_50561119 | 5.03 |
ENST00000555216.5
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr7_-_22193824 | 4.45 |
ENST00000401957.6
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr4_-_158159657 | 4.28 |
ENST00000590648.5
|
GASK1B
|
golgi associated kinase 1B |
| chr16_-_58546702 | 4.14 |
ENST00000567133.1
|
CNOT1
|
CCR4-NOT transcription complex subunit 1 |
| chr9_-_120714457 | 4.14 |
ENST00000373930.4
|
MEGF9
|
multiple EGF like domains 9 |
| chr3_-_71305986 | 4.00 |
ENST00000647614.1
|
FOXP1
|
forkhead box P1 |
| chr8_-_17722217 | 3.88 |
ENST00000381861.7
|
MTUS1
|
microtubule associated scaffold protein 1 |
| chr4_+_41538143 | 3.73 |
ENST00000503057.6
ENST00000511496.5 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr2_+_69013170 | 3.63 |
ENST00000303714.9
|
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr7_+_16753731 | 3.63 |
ENST00000262067.5
|
TSPAN13
|
tetraspanin 13 |
| chr8_-_80080816 | 3.55 |
ENST00000520527.5
ENST00000517427.5 ENST00000379097.7 ENST00000448733.3 |
TPD52
|
tumor protein D52 |
| chr3_-_52685794 | 3.54 |
ENST00000424867.1
ENST00000394830.7 ENST00000431678.5 ENST00000450271.5 |
PBRM1
|
polybromo 1 |
| chr4_-_22515932 | 3.37 |
ENST00000502482.1
|
ADGRA3
|
adhesion G protein-coupled receptor A3 |
| chr15_-_82045998 | 3.15 |
ENST00000329713.5
|
MEX3B
|
mex-3 RNA binding family member B |
| chr18_+_6729698 | 3.10 |
ENST00000383472.9
|
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr2_-_37672178 | 3.08 |
ENST00000457889.1
|
CDC42EP3
|
CDC42 effector protein 3 |
| chr15_-_82046119 | 3.08 |
ENST00000558133.1
|
MEX3B
|
mex-3 RNA binding family member B |
| chr5_-_111756245 | 3.07 |
ENST00000447165.6
|
NREP
|
neuronal regeneration related protein |
| chr17_-_44199206 | 3.03 |
ENST00000589805.1
|
ATXN7L3
|
ataxin 7 like 3 |
| chr7_+_102912983 | 2.95 |
ENST00000339431.9
ENST00000249377.4 |
LRRC17
|
leucine rich repeat containing 17 |
| chr5_-_111758061 | 2.95 |
ENST00000509979.5
ENST00000513100.5 ENST00000508161.5 ENST00000455559.6 |
NREP
|
neuronal regeneration related protein |
| chr1_+_185734362 | 2.92 |
ENST00000271588.9
|
HMCN1
|
hemicentin 1 |
| chr4_+_54657918 | 2.89 |
ENST00000412167.6
ENST00000288135.6 |
KIT
|
KIT proto-oncogene, receptor tyrosine kinase |
| chr16_+_30395400 | 2.84 |
ENST00000320159.2
ENST00000613509.2 |
ZNF48
|
zinc finger protein 48 |
| chr1_-_46132650 | 2.81 |
ENST00000372006.5
ENST00000425892.2 ENST00000420542.5 |
PIK3R3
|
phosphoinositide-3-kinase regulatory subunit 3 |
| chr13_-_106535653 | 2.75 |
ENST00000646441.1
|
EFNB2
|
ephrin B2 |
| chrX_-_117973579 | 2.73 |
ENST00000371878.5
|
KLHL13
|
kelch like family member 13 |
| chr2_-_136116165 | 2.72 |
ENST00000409817.1
|
CXCR4
|
C-X-C motif chemokine receptor 4 |
| chr3_-_18424533 | 2.67 |
ENST00000417717.6
|
SATB1
|
SATB homeobox 1 |
| chr5_+_122312229 | 2.65 |
ENST00000261368.13
|
SNCAIP
|
synuclein alpha interacting protein |
| chr5_+_122312164 | 2.61 |
ENST00000514497.6
ENST00000261367.11 |
SNCAIP
|
synuclein alpha interacting protein |
| chr6_+_30880780 | 2.55 |
ENST00000460944.6
ENST00000324771.12 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr14_-_87993143 | 2.52 |
ENST00000622264.4
|
GALC
|
galactosylceramidase |
| chr21_-_26573211 | 2.51 |
ENST00000299340.9
ENST00000652641.2 |
CYYR1
|
cysteine and tyrosine rich 1 |
| chr5_-_111757704 | 2.49 |
ENST00000379671.7
|
NREP
|
neuronal regeneration related protein |
| chr16_+_67029359 | 2.45 |
ENST00000565389.1
|
CBFB
|
core-binding factor subunit beta |
| chr9_-_72364504 | 2.44 |
ENST00000237937.7
ENST00000343431.6 ENST00000376956.3 |
ZFAND5
|
zinc finger AN1-type containing 5 |
| chr9_+_88388356 | 2.43 |
ENST00000375859.4
|
SPIN1
|
spindlin 1 |
| chr1_+_28914597 | 2.42 |
ENST00000349460.9
|
EPB41
|
erythrocyte membrane protein band 4.1 |
| chr6_+_125919296 | 2.37 |
ENST00000444128.2
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr6_+_125919210 | 2.34 |
ENST00000438495.6
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr13_-_100674774 | 2.30 |
ENST00000328767.9
|
TMTC4
|
transmembrane O-mannosyltransferase targeting cadherins 4 |
| chrX_+_15500800 | 2.25 |
ENST00000348343.11
|
BMX
|
BMX non-receptor tyrosine kinase |
| chr13_+_111115303 | 2.24 |
ENST00000646102.2
ENST00000449979.5 |
ARHGEF7
|
Rho guanine nucleotide exchange factor 7 |
| chr17_-_64390852 | 2.23 |
ENST00000563924.6
|
PECAM1
|
platelet and endothelial cell adhesion molecule 1 |
| chr11_-_10808304 | 2.21 |
ENST00000532082.6
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma 2 |
| chr12_+_93378625 | 2.20 |
ENST00000546925.1
|
NUDT4
|
nudix hydrolase 4 |
| chr4_+_145482761 | 2.19 |
ENST00000507367.1
ENST00000394092.6 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr1_+_182839338 | 2.19 |
ENST00000367549.4
|
DHX9
|
DExH-box helicase 9 |
| chr8_-_6563044 | 2.18 |
ENST00000338312.10
|
ANGPT2
|
angiopoietin 2 |
| chr1_-_25906457 | 2.16 |
ENST00000426559.6
|
STMN1
|
stathmin 1 |
| chrX_+_85244075 | 2.15 |
ENST00000276123.7
|
ZNF711
|
zinc finger protein 711 |
| chr22_-_35961623 | 2.14 |
ENST00000408983.2
|
RBFOX2
|
RNA binding fox-1 homolog 2 |
| chr14_+_96039328 | 2.05 |
ENST00000553764.1
ENST00000555004.3 ENST00000556728.1 ENST00000553782.1 |
C14orf132
|
chromosome 14 open reading frame 132 |
| chr10_+_6202866 | 2.03 |
ENST00000317350.8
ENST00000379785.5 ENST00000625260.2 ENST00000626882.2 ENST00000360521.7 ENST00000379775.9 ENST00000640683.1 |
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr12_-_6124662 | 2.02 |
ENST00000261405.10
|
VWF
|
von Willebrand factor |
| chr9_-_71768386 | 2.02 |
ENST00000377066.9
ENST00000377044.9 |
CEMIP2
|
cell migration inducing hyaluronidase 2 |
| chr10_+_18659382 | 2.00 |
ENST00000377275.4
|
ARL5B
|
ADP ribosylation factor like GTPase 5B |
| chr7_+_77538027 | 2.00 |
ENST00000433369.6
ENST00000415482.6 |
PTPN12
|
protein tyrosine phosphatase non-receptor type 12 |
| chr1_+_28914614 | 1.98 |
ENST00000645184.1
|
EPB41
|
erythrocyte membrane protein band 4.1 |
| chr16_+_67029093 | 1.97 |
ENST00000561924.6
|
CBFB
|
core-binding factor subunit beta |
| chr1_+_164559739 | 1.97 |
ENST00000627490.2
|
PBX1
|
PBX homeobox 1 |
| chr1_+_47023659 | 1.96 |
ENST00000371901.4
|
CYP4X1
|
cytochrome P450 family 4 subfamily X member 1 |
| chr7_+_116672187 | 1.95 |
ENST00000318493.11
ENST00000397752.8 |
MET
|
MET proto-oncogene, receptor tyrosine kinase |
| chr1_+_84181630 | 1.94 |
ENST00000610457.1
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr7_-_32892015 | 1.93 |
ENST00000452926.1
|
KBTBD2
|
kelch repeat and BTB domain containing 2 |
| chr16_+_11668414 | 1.93 |
ENST00000329565.6
|
SNN
|
stannin |
| chr1_-_182391363 | 1.93 |
ENST00000417584.6
|
GLUL
|
glutamate-ammonia ligase |
| chr15_-_70096604 | 1.86 |
ENST00000559048.5
ENST00000560939.5 ENST00000440567.7 ENST00000557907.5 ENST00000558379.5 ENST00000559929.5 |
TLE3
|
TLE family member 3, transcriptional corepressor |
| chr17_-_63699730 | 1.84 |
ENST00000578061.5
|
LIMD2
|
LIM domain containing 2 |
| chr20_-_4823597 | 1.84 |
ENST00000379400.8
|
RASSF2
|
Ras association domain family member 2 |
| chr1_+_65992389 | 1.83 |
ENST00000423207.6
|
PDE4B
|
phosphodiesterase 4B |
| chr7_-_106285898 | 1.83 |
ENST00000424768.2
ENST00000681255.1 |
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr19_-_18941117 | 1.77 |
ENST00000600077.5
|
HOMER3
|
homer scaffold protein 3 |
| chr3_+_19947316 | 1.74 |
ENST00000422242.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr9_+_87498491 | 1.73 |
ENST00000622514.4
|
DAPK1
|
death associated protein kinase 1 |
| chr4_+_56049090 | 1.73 |
ENST00000629263.3
ENST00000682029.1 |
CRACD
|
capping protein inhibiting regulator of actin dynamics |
| chr4_+_77158252 | 1.73 |
ENST00000395640.5
|
CCNG2
|
cyclin G2 |
| chr5_-_1112051 | 1.73 |
ENST00000264930.10
|
SLC12A7
|
solute carrier family 12 member 7 |
| chr6_+_79631322 | 1.70 |
ENST00000369838.6
|
SH3BGRL2
|
SH3 domain binding glutamate rich protein like 2 |
| chr16_-_46831043 | 1.69 |
ENST00000565112.1
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr14_-_34874887 | 1.69 |
ENST00000382422.6
|
BAZ1A
|
bromodomain adjacent to zinc finger domain 1A |
| chr12_+_93378550 | 1.69 |
ENST00000550056.5
ENST00000549992.5 ENST00000548662.5 ENST00000547014.5 |
NUDT4
|
nudix hydrolase 4 |
| chr3_-_142028597 | 1.68 |
ENST00000467667.5
|
TFDP2
|
transcription factor Dp-2 |
| chr7_-_22194709 | 1.67 |
ENST00000458533.5
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr4_+_155758990 | 1.67 |
ENST00000505154.5
ENST00000652626.1 ENST00000502959.5 ENST00000264424.13 ENST00000505764.5 ENST00000507146.5 ENST00000503520.5 |
GUCY1B1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr10_-_73591330 | 1.66 |
ENST00000451492.5
ENST00000681793.1 ENST00000680396.1 ENST00000413442.5 |
USP54
|
ubiquitin specific peptidase 54 |
| chr2_-_207624551 | 1.66 |
ENST00000272839.7
ENST00000426075.5 |
METTL21A
|
methyltransferase like 21A |
| chr6_-_11232658 | 1.66 |
ENST00000379433.5
ENST00000379446.10 ENST00000620854.4 |
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chrX_+_85244032 | 1.66 |
ENST00000373165.7
|
ZNF711
|
zinc finger protein 711 |
| chr14_-_87992838 | 1.65 |
ENST00000544807.6
|
GALC
|
galactosylceramidase |
| chr2_-_207167220 | 1.64 |
ENST00000421199.5
ENST00000457962.5 |
KLF7
|
Kruppel like factor 7 |
| chr4_-_140756315 | 1.64 |
ENST00000442267.3
|
TBC1D9
|
TBC1 domain family member 9 |
| chr4_-_185810894 | 1.63 |
ENST00000448662.6
ENST00000439049.5 ENST00000420158.5 ENST00000319471.13 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr10_+_21524627 | 1.63 |
ENST00000651097.1
|
MLLT10
|
MLLT10 histone lysine methyltransferase DOT1L cofactor |
| chr4_+_77158418 | 1.63 |
ENST00000509972.1
|
CCNG2
|
cyclin G2 |
| chr5_-_111757382 | 1.63 |
ENST00000453526.6
ENST00000509427.5 |
NREP
|
neuronal regeneration related protein |
| chrX_+_85243983 | 1.63 |
ENST00000674551.1
|
ZNF711
|
zinc finger protein 711 |
| chr15_-_52652031 | 1.63 |
ENST00000546305.6
|
FAM214A
|
family with sequence similarity 214 member A |
| chrX_-_134915203 | 1.62 |
ENST00000370779.8
|
MOSPD1
|
motile sperm domain containing 1 |
| chr19_+_39413528 | 1.61 |
ENST00000438123.5
ENST00000409797.6 ENST00000451354.6 |
PLEKHG2
|
pleckstrin homology and RhoGEF domain containing G2 |
| chr5_-_111757549 | 1.61 |
ENST00000419114.6
|
NREP
|
neuronal regeneration related protein |
| chr4_+_143336762 | 1.59 |
ENST00000262995.8
|
GAB1
|
GRB2 associated binding protein 1 |
| chr20_-_45348414 | 1.58 |
ENST00000372733.3
|
SDC4
|
syndecan 4 |
| chr13_+_32031300 | 1.57 |
ENST00000642040.1
|
FRY
|
FRY microtubule binding protein |
| chr2_-_237590660 | 1.57 |
ENST00000409576.1
|
RAB17
|
RAB17, member RAS oncogene family |
| chr2_+_111122631 | 1.56 |
ENST00000393252.3
|
BCL2L11
|
BCL2 like 11 |
| chr5_-_111757465 | 1.56 |
ENST00000446294.6
|
NREP
|
neuronal regeneration related protein |
| chr9_+_79572715 | 1.56 |
ENST00000265284.10
|
TLE4
|
TLE family member 4, transcriptional corepressor |
| chr20_-_20712626 | 1.55 |
ENST00000202677.12
|
RALGAPA2
|
Ral GTPase activating protein catalytic subunit alpha 2 |
| chr14_-_87993159 | 1.55 |
ENST00000393568.8
ENST00000261304.7 |
GALC
|
galactosylceramidase |
| chr5_+_141417659 | 1.55 |
ENST00000398594.4
|
PCDHGB7
|
protocadherin gamma subfamily B, 7 |
| chr9_+_79572572 | 1.55 |
ENST00000435650.5
ENST00000414465.5 ENST00000376537.8 |
TLE4
|
TLE family member 4, transcriptional corepressor |
| chr21_-_26051023 | 1.55 |
ENST00000415997.1
|
APP
|
amyloid beta precursor protein |
| chr14_-_34875348 | 1.53 |
ENST00000360310.6
|
BAZ1A
|
bromodomain adjacent to zinc finger domain 1A |
| chr19_-_49361475 | 1.53 |
ENST00000598810.5
|
TEAD2
|
TEA domain transcription factor 2 |
| chr1_+_60865259 | 1.53 |
ENST00000371191.5
|
NFIA
|
nuclear factor I A |
| chr6_-_75206044 | 1.53 |
ENST00000322507.13
|
COL12A1
|
collagen type XII alpha 1 chain |
| chr1_+_32765667 | 1.53 |
ENST00000373480.1
|
KIAA1522
|
KIAA1522 |
| chr8_-_38467701 | 1.50 |
ENST00000425967.8
ENST00000533668.5 ENST00000413133.6 ENST00000397108.8 ENST00000526742.5 ENST00000525001.5 ENST00000529552.5 ENST00000397113.6 |
FGFR1
|
fibroblast growth factor receptor 1 |
| chr6_+_154995258 | 1.50 |
ENST00000682666.1
|
TIAM2
|
TIAM Rac1 associated GEF 2 |
| chr1_+_40450053 | 1.49 |
ENST00000484445.5
ENST00000411995.6 |
ZFP69B
|
ZFP69 zinc finger protein B |
| chr15_-_50355100 | 1.48 |
ENST00000543881.5
|
GABPB1
|
GA binding protein transcription factor subunit beta 1 |
| chr3_-_126101506 | 1.48 |
ENST00000509064.5
ENST00000508835.5 |
SLC41A3
|
solute carrier family 41 member 3 |
| chr14_+_103123452 | 1.48 |
ENST00000558056.1
ENST00000560869.6 |
TNFAIP2
|
TNF alpha induced protein 2 |
| chr5_+_141421064 | 1.48 |
ENST00000518882.2
|
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr2_+_12716893 | 1.47 |
ENST00000381465.2
ENST00000155926.9 |
TRIB2
|
tribbles pseudokinase 2 |
| chr8_-_6563238 | 1.46 |
ENST00000629816.3
ENST00000523120.2 |
ANGPT2
|
angiopoietin 2 |
| chr3_-_27484374 | 1.46 |
ENST00000445684.5
ENST00000425128.6 |
SLC4A7
|
solute carrier family 4 member 7 |
| chrX_-_45200895 | 1.46 |
ENST00000377934.4
|
DIPK2B
|
divergent protein kinase domain 2B |
| chr3_+_183265302 | 1.45 |
ENST00000465010.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr3_+_152268920 | 1.44 |
ENST00000495875.6
ENST00000324210.10 ENST00000493459.5 |
MBNL1
|
muscleblind like splicing regulator 1 |
| chr15_-_59372863 | 1.44 |
ENST00000288235.9
|
MYO1E
|
myosin IE |
| chr8_+_141128612 | 1.43 |
ENST00000518347.5
ENST00000262585.6 ENST00000520986.5 ENST00000523058.5 ENST00000518668.5 |
DENND3
|
DENN domain containing 3 |
| chr2_-_207166818 | 1.42 |
ENST00000423015.5
|
KLF7
|
Kruppel like factor 7 |
| chr11_-_66677748 | 1.41 |
ENST00000525754.5
ENST00000531969.5 ENST00000524637.1 ENST00000531036.2 ENST00000310046.9 |
RBM4B
|
RNA binding motif protein 4B |
| chr1_-_201154418 | 1.41 |
ENST00000435310.5
ENST00000485839.6 |
TMEM9
|
transmembrane protein 9 |
| chr13_-_100674787 | 1.40 |
ENST00000342624.10
|
TMTC4
|
transmembrane O-mannosyltransferase targeting cadherins 4 |
| chr2_-_25982471 | 1.39 |
ENST00000264712.8
|
KIF3C
|
kinesin family member 3C |
| chr8_-_6563409 | 1.39 |
ENST00000325203.9
|
ANGPT2
|
angiopoietin 2 |
| chr17_+_67377413 | 1.38 |
ENST00000580974.5
|
PITPNC1
|
phosphatidylinositol transfer protein cytoplasmic 1 |
| chr20_+_6767678 | 1.38 |
ENST00000378827.5
|
BMP2
|
bone morphogenetic protein 2 |
| chr4_-_89836213 | 1.37 |
ENST00000618500.4
ENST00000508895.5 |
SNCA
|
synuclein alpha |
| chr4_-_185956348 | 1.36 |
ENST00000431902.5
ENST00000284776.11 ENST00000415274.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr17_+_67377272 | 1.35 |
ENST00000581322.6
ENST00000299954.13 |
PITPNC1
|
phosphatidylinositol transfer protein cytoplasmic 1 |
| chr3_-_71306012 | 1.34 |
ENST00000649431.1
ENST00000610810.5 |
FOXP1
|
forkhead box P1 |
| chr7_+_74964692 | 1.34 |
ENST00000616305.2
|
CASTOR2
|
cytosolic arginine sensor for mTORC1 subunit 2 |
| chr4_-_89835617 | 1.34 |
ENST00000611107.1
ENST00000345009.8 ENST00000505199.5 ENST00000502987.5 |
SNCA
|
synuclein alpha |
| chr4_+_77157189 | 1.33 |
ENST00000316355.10
ENST00000502280.5 |
CCNG2
|
cyclin G2 |
| chr6_-_35688907 | 1.33 |
ENST00000539068.5
ENST00000357266.9 |
FKBP5
|
FKBP prolyl isomerase 5 |
| chr18_+_3448456 | 1.33 |
ENST00000549780.5
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr4_+_143336868 | 1.32 |
ENST00000262994.8
|
GAB1
|
GRB2 associated binding protein 1 |
| chr3_+_141324208 | 1.32 |
ENST00000509842.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr3_-_142029108 | 1.32 |
ENST00000497579.5
|
TFDP2
|
transcription factor Dp-2 |
| chr7_-_106285094 | 1.32 |
ENST00000680584.1
ENST00000354289.9 |
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr5_-_172283743 | 1.32 |
ENST00000393792.3
|
UBTD2
|
ubiquitin domain containing 2 |
| chr14_-_91946989 | 1.31 |
ENST00000556154.5
|
FBLN5
|
fibulin 5 |
| chr1_-_235328147 | 1.31 |
ENST00000264183.9
ENST00000418304.1 ENST00000349213.7 |
ARID4B
|
AT-rich interaction domain 4B |
| chr11_+_125164743 | 1.31 |
ENST00000298282.14
|
PKNOX2
|
PBX/knotted 1 homeobox 2 |
| chr5_-_116574802 | 1.30 |
ENST00000343348.11
|
SEMA6A
|
semaphorin 6A |
| chr17_-_81870974 | 1.29 |
ENST00000541078.6
|
ARHGDIA
|
Rho GDP dissociation inhibitor alpha |
| chr15_-_72783611 | 1.29 |
ENST00000563907.5
|
ADPGK
|
ADP dependent glucokinase |
| chr9_-_14307928 | 1.28 |
ENST00000637640.1
ENST00000493697.1 ENST00000636057.1 |
NFIB
|
nuclear factor I B |
| chr3_-_27484335 | 1.28 |
ENST00000454389.5
ENST00000440156.5 ENST00000437179.5 ENST00000446700.5 ENST00000455077.5 |
SLC4A7
|
solute carrier family 4 member 7 |
| chr4_+_153466324 | 1.28 |
ENST00000409663.7
ENST00000409959.8 |
TMEM131L
|
transmembrane 131 like |
| chrX_+_81202066 | 1.27 |
ENST00000373212.6
|
SH3BGRL
|
SH3 domain binding glutamate rich protein like |
| chr22_-_37484505 | 1.27 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr6_-_129710145 | 1.26 |
ENST00000368149.3
|
ARHGAP18
|
Rho GTPase activating protein 18 |
| chr8_-_8893548 | 1.26 |
ENST00000276282.7
|
MFHAS1
|
malignant fibrous histiocytoma amplified sequence 1 |
| chr9_-_72365198 | 1.25 |
ENST00000376962.10
ENST00000376960.8 |
ZFAND5
|
zinc finger AN1-type containing 5 |
| chr7_+_116210501 | 1.25 |
ENST00000455989.1
ENST00000358204.9 |
TES
|
testin LIM domain protein |
| chr11_+_94128834 | 1.24 |
ENST00000227638.8
ENST00000436171.2 |
PANX1
|
pannexin 1 |
| chr12_-_31324129 | 1.24 |
ENST00000454658.6
|
SINHCAF
|
SIN3-HDAC complex associated factor |
| chr3_-_65597886 | 1.23 |
ENST00000460329.6
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr2_+_176151543 | 1.22 |
ENST00000306324.4
|
HOXD4
|
homeobox D4 |
| chr14_-_50231570 | 1.22 |
ENST00000216373.10
|
SOS2
|
SOS Ras/Rho guanine nucleotide exchange factor 2 |
| chr9_+_131096476 | 1.22 |
ENST00000372309.7
ENST00000247291.8 ENST00000372302.5 ENST00000372300.5 ENST00000372298.1 |
AIF1L
|
allograft inflammatory factor 1 like |
| chrX_-_45200828 | 1.22 |
ENST00000398000.7
|
DIPK2B
|
divergent protein kinase domain 2B |
| chr4_-_22516001 | 1.20 |
ENST00000334304.10
|
ADGRA3
|
adhesion G protein-coupled receptor A3 |
| chr8_-_38468627 | 1.20 |
ENST00000683815.1
ENST00000684654.1 ENST00000447712.7 ENST00000397091.9 |
FGFR1
|
fibroblast growth factor receptor 1 |
| chr3_+_183635605 | 1.19 |
ENST00000493074.1
ENST00000437402.5 ENST00000454495.6 ENST00000473045.5 ENST00000468101.5 ENST00000427201.2 ENST00000482138.1 ENST00000454652.6 ENST00000242810.11 |
KLHL24
|
kelch like family member 24 |
| chr1_+_65420643 | 1.19 |
ENST00000371059.7
ENST00000371060.7 ENST00000349533.11 ENST00000371065.9 ENST00000488747.5 ENST00000484243.1 ENST00000613538.1 |
LEPR
LEPROT
|
leptin receptor leptin receptor overlapping transcript |
| chr8_-_38468601 | 1.19 |
ENST00000341462.9
ENST00000683765.1 ENST00000356207.9 ENST00000326324.10 ENST00000335922.9 ENST00000532791.5 |
FGFR1
|
fibroblast growth factor receptor 1 |
| chr1_-_21937300 | 1.19 |
ENST00000374695.8
|
HSPG2
|
heparan sulfate proteoglycan 2 |
| chr7_-_11832190 | 1.19 |
ENST00000423059.9
ENST00000617773.1 |
THSD7A
|
thrombospondin type 1 domain containing 7A |
| chr1_-_32336224 | 1.18 |
ENST00000329421.8
|
MARCKSL1
|
MARCKS like 1 |
| chr22_-_32255344 | 1.18 |
ENST00000266086.6
|
SLC5A4
|
solute carrier family 5 member 4 |
| chr10_-_89414458 | 1.18 |
ENST00000371837.5
|
LIPA
|
lipase A, lysosomal acid type |
| chr8_+_141417902 | 1.17 |
ENST00000681443.1
|
PTP4A3
|
protein tyrosine phosphatase 4A3 |
| chr19_-_18941184 | 1.17 |
ENST00000594794.5
ENST00000392351.8 ENST00000596482.5 |
HOMER3
|
homer scaffold protein 3 |
| chr6_+_44247866 | 1.16 |
ENST00000371554.2
|
HSP90AB1
|
heat shock protein 90 alpha family class B member 1 |
| chr5_+_149581368 | 1.16 |
ENST00000333677.7
|
ARHGEF37
|
Rho guanine nucleotide exchange factor 37 |
| chr16_+_2033264 | 1.16 |
ENST00000565855.5
ENST00000566198.1 |
SLC9A3R2
|
SLC9A3 regulator 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0006683 | galactosylceramide catabolic process(GO:0006683) |
| 1.7 | 5.0 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 1.3 | 3.9 | GO:1903465 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 1.1 | 3.3 | GO:0070662 | mast cell proliferation(GO:0070662) |
| 1.0 | 4.8 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.7 | 2.8 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.7 | 7.9 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.6 | 2.6 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.6 | 3.9 | GO:1901907 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.6 | 1.8 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 0.6 | 2.2 | GO:0072011 | diapedesis(GO:0050904) glomerular endothelium development(GO:0072011) |
| 0.5 | 1.6 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.5 | 1.6 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.5 | 1.5 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.5 | 1.5 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.5 | 8.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.5 | 1.4 | GO:0060128 | regulation of calcium-independent cell-cell adhesion(GO:0051040) corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.5 | 0.9 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.5 | 1.4 | GO:0097252 | oligodendrocyte apoptotic process(GO:0097252) |
| 0.4 | 4.0 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.4 | 1.3 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.4 | 1.3 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.4 | 1.2 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.4 | 5.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.4 | 0.8 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.4 | 5.7 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.4 | 2.7 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.4 | 1.2 | GO:1901389 | regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 0.4 | 1.1 | GO:0099557 | trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission(GO:0099557) |
| 0.4 | 3.3 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.4 | 1.5 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.4 | 2.2 | GO:0035549 | positive regulation of interferon-beta secretion(GO:0035549) |
| 0.4 | 1.5 | GO:2000329 | negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.3 | 0.7 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.3 | 2.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.3 | 2.8 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.3 | 0.9 | GO:0009720 | detection of hormone stimulus(GO:0009720) |
| 0.3 | 0.9 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.3 | 2.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.3 | 2.6 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.3 | 0.9 | GO:2000793 | cell proliferation involved in heart valve development(GO:2000793) |
| 0.3 | 1.1 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.3 | 5.0 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.3 | 0.6 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.3 | 0.8 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.3 | 1.6 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.3 | 0.8 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.3 | 3.2 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.3 | 1.3 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.3 | 1.5 | GO:0002265 | astrocyte activation involved in immune response(GO:0002265) microglia differentiation(GO:0014004) microglia development(GO:0014005) collateral sprouting in absence of injury(GO:0048669) smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.3 | 0.8 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.2 | 1.2 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.2 | 0.7 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.2 | 0.7 | GO:0044335 | canonical Wnt signaling pathway involved in neural crest cell differentiation(GO:0044335) |
| 0.2 | 4.8 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.2 | 1.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.2 | 2.5 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 | 8.0 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.2 | 0.9 | GO:0043387 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.2 | 1.5 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.2 | 0.2 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.2 | 1.3 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.2 | 2.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.2 | 1.9 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.2 | 0.6 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.2 | 0.4 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.2 | 3.3 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.2 | 0.4 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.2 | 1.1 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.2 | 0.9 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.2 | 0.9 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.2 | 3.5 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.2 | 0.6 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.2 | 1.3 | GO:0030421 | defecation(GO:0030421) |
| 0.2 | 0.2 | GO:0035787 | cell migration involved in kidney development(GO:0035787) |
| 0.2 | 1.4 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.2 | 0.5 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.2 | 3.0 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.2 | 0.9 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.2 | 0.7 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.2 | 2.9 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 | 0.5 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.2 | 2.0 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.2 | 0.5 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.2 | 2.6 | GO:1903764 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.2 | 0.5 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.2 | 1.3 | GO:0071352 | cellular response to interleukin-2(GO:0071352) |
| 0.2 | 1.7 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.2 | 1.6 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.2 | 1.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.2 | 2.3 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.2 | 5.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 0.5 | GO:0061027 | umbilical cord morphogenesis(GO:0036304) umbilical cord development(GO:0061027) |
| 0.1 | 3.6 | GO:2000121 | regulation of removal of superoxide radicals(GO:2000121) |
| 0.1 | 0.1 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 0.9 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 3.2 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.1 | 0.6 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 0.7 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.7 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.1 | 0.6 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.1 | 0.9 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.1 | 1.1 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.3 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.1 | 1.3 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 | 0.4 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.8 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.1 | 0.6 | GO:0090156 | negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.1 | 0.4 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) |
| 0.1 | 0.7 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 5.0 | GO:2000810 | regulation of bicellular tight junction assembly(GO:2000810) |
| 0.1 | 2.4 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 0.6 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 | 0.3 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.1 | 0.3 | GO:0006113 | fermentation(GO:0006113) regulation of fermentation(GO:0043465) |
| 0.1 | 0.4 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.1 | 2.1 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) |
| 0.1 | 0.3 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.1 | 0.9 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 1.5 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.7 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 1.8 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 1.0 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.6 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.3 | GO:1904247 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 0.3 | GO:0001828 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.1 | 0.9 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.1 | 0.4 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.4 | GO:0018032 | protein amidation(GO:0018032) |
| 0.1 | 3.6 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.1 | 1.4 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 0.3 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 2.5 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.3 | GO:0046709 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.1 | 0.4 | GO:1990834 | response to odorant(GO:1990834) |
| 0.1 | 0.7 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 0.4 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.1 | 0.5 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.1 | 0.3 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.3 | GO:0036484 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.1 | 0.6 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.1 | 2.7 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 0.2 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.1 | 0.6 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.1 | 0.8 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.1 | 4.3 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 0.5 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.5 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 3.8 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.4 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.3 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.1 | 0.4 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.1 | 0.4 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.4 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 0.4 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 | 0.9 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 0.2 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.1 | 0.8 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.2 | GO:1903551 | extracellular exosome assembly(GO:0071971) regulation of extracellular exosome assembly(GO:1903551) |
| 0.1 | 0.8 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 0.3 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.1 | 1.6 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 0.5 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 0.2 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 1.8 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.1 | 1.8 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 0.3 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.1 | 1.2 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.1 | 0.8 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.1 | 1.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.3 | GO:0003290 | atrial septum secundum morphogenesis(GO:0003290) |
| 0.1 | 0.4 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.1 | 0.2 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.1 | 1.0 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.2 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 2.5 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 1.0 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.9 | GO:0044334 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.1 | 0.4 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.1 | 0.2 | GO:0097698 | telomere maintenance via base-excision repair(GO:0097698) |
| 0.1 | 0.3 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 0.8 | GO:0071073 | positive regulation of phospholipid biosynthetic process(GO:0071073) |
| 0.1 | 1.8 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.5 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 2.3 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.7 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 1.7 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.4 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 | 0.2 | GO:0075732 | viral penetration into host nucleus(GO:0075732) multi-organism nuclear import(GO:1902594) |
| 0.1 | 2.1 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 2.3 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 1.1 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.4 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.4 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.1 | 3.9 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.1 | 0.4 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 2.1 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.1 | 0.4 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.4 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 1.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 0.2 | GO:2000642 | intralumenal vesicle formation(GO:0070676) negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 | 0.5 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 0.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 0.2 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.1 | 1.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 4.8 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.1 | 0.2 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 | 0.9 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.1 | 0.6 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.2 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 2.1 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.1 | 0.9 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.1 | 0.6 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.5 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.1 | 0.4 | GO:0072161 | mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.1 | 0.2 | GO:0099403 | negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 0.1 | 0.9 | GO:0050812 | regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.1 | 0.5 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.2 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.1 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.0 | 0.4 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.2 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.3 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 1.7 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.2 | GO:1903755 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.0 | 0.9 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.2 | GO:1905049 | negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 | 0.7 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.3 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.6 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0021965 | spinal cord ventral commissure morphogenesis(GO:0021965) dorsal/ventral axon guidance(GO:0033563) |
| 0.0 | 13.0 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.9 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 9.5 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.2 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.7 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.3 | GO:0060260 | regulation of transcription initiation from RNA polymerase II promoter(GO:0060260) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 1.1 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.3 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.6 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.8 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.3 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 1.8 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.3 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.8 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.0 | GO:0060268 | negative regulation of respiratory burst(GO:0060268) |
| 0.0 | 0.1 | GO:0070894 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 0.0 | 0.1 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 | 0.3 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.3 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.9 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.2 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.3 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.5 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.7 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 1.0 | GO:0061377 | mammary gland alveolus development(GO:0060749) mammary gland lobule development(GO:0061377) |
| 0.0 | 0.2 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 2.0 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.3 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.2 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.0 | 1.0 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.8 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.2 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.1 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.0 | 3.7 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 1.3 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.3 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.3 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.4 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:2000174 | pro-T cell differentiation(GO:0002572) regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.0 | 0.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.3 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.0 | GO:2000211 | negative regulation of cellular amino acid metabolic process(GO:0045763) regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.4 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:1902161 | transepithelial water transport(GO:0035377) positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.1 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.2 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 2.7 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.3 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.2 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.7 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.4 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.2 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.3 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.2 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.0 | 0.2 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.3 | GO:0034391 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 | 0.9 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.8 | GO:0006768 | biotin metabolic process(GO:0006768) |
| 0.0 | 0.1 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.8 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 0.1 | GO:0052031 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.0 | 0.5 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.0 | 0.3 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 3.9 | GO:0006275 | regulation of DNA replication(GO:0006275) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.2 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.5 | GO:0006527 | citrulline metabolic process(GO:0000052) arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.8 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.6 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 1.7 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.3 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.3 | GO:0006547 | histidine metabolic process(GO:0006547) |
| 0.0 | 0.3 | GO:2000425 | regulation of apoptotic cell clearance(GO:2000425) |
| 0.0 | 1.5 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.0 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.5 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.2 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.0 | 0.2 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.4 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.2 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 3.0 | GO:0009267 | cellular response to starvation(GO:0009267) |
| 0.0 | 0.2 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.1 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.4 | GO:0051354 | negative regulation of oxidoreductase activity(GO:0051354) |
| 0.0 | 0.4 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.7 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.6 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.0 | 0.1 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.9 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.2 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.1 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.0 | 3.0 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.7 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.6 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.5 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.3 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.3 | GO:0036109 | alpha-linolenic acid metabolic process(GO:0036109) |
| 0.0 | 0.4 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.2 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.1 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:1902430 | negative regulation of beta-amyloid formation(GO:1902430) |
| 0.0 | 0.5 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 3.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.3 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.7 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.5 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 0.5 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.4 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 0.1 | GO:0031437 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of IRE1-mediated unfolded protein response(GO:1903895) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.2 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.5 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.2 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.2 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.0 | 0.0 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.5 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.4 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.5 | GO:1903170 | negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.0 | 3.6 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 1.0 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 | 0.0 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.1 | GO:0002774 | Fc receptor mediated inhibitory signaling pathway(GO:0002774) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.0 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.4 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 1.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.4 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.0 | 0.0 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.9 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 0.8 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 0.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.0 | 0.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.3 | GO:0003179 | heart valve morphogenesis(GO:0003179) |
| 0.0 | 0.2 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.4 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.3 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 1.4 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 0.2 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 8.2 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 0.0 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.3 | GO:0045840 | positive regulation of mitotic nuclear division(GO:0045840) |
| 0.0 | 0.2 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.5 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.0 | GO:0010751 | regulation of nitric oxide mediated signal transduction(GO:0010749) negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.0 | 0.2 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.4 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.4 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.2 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.3 | GO:0042594 | response to starvation(GO:0042594) |
| 0.0 | 0.3 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 2.0 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0008623 | CHRAC(GO:0008623) |
| 0.6 | 2.4 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.6 | 1.8 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.5 | 2.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.5 | 3.6 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.5 | 3.7 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.4 | 0.4 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.4 | 1.6 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.4 | 1.2 | GO:1990913 | sperm head plasma membrane(GO:1990913) ooplasm(GO:1990917) |
| 0.4 | 2.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.3 | 1.3 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.3 | 1.9 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.3 | 2.9 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.3 | 4.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.3 | 3.8 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.2 | 2.6 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.2 | 1.8 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.2 | 0.6 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.2 | 4.7 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.2 | 4.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.2 | 7.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.2 | 3.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.2 | 0.9 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.2 | 2.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.2 | 0.8 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.2 | 2.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 1.7 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.2 | 0.9 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 2.1 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 0.9 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 5.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 1.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 2.6 | GO:0043219 | lateral loop(GO:0043219) |
| 0.1 | 0.8 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 2.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.4 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) integrin alpha7-beta1 complex(GO:0034677) |
| 0.1 | 0.6 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 2.1 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 3.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 2.7 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 1.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.6 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.7 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 0.5 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 0.7 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 2.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.5 | GO:0035363 | histone locus body(GO:0035363) |
| 0.1 | 0.5 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 2.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 15.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 1.1 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 0.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 1.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.7 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 3.4 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 1.3 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.1 | 0.4 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 0.9 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 1.6 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 7.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 2.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 1.2 | GO:0032059 | bleb(GO:0032059) |
| 0.1 | 0.9 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 0.6 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 0.6 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.7 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 0.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.8 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 0.6 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 0.6 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.4 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 0.2 | GO:0072517 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.1 | 4.7 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 0.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.7 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:1990356 | sumoylated E2 ligase complex(GO:1990356) |
| 0.0 | 0.9 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 1.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.9 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.4 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.6 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 1.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 7.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 2.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.9 | GO:1990752 | microtubule end(GO:1990752) |
| 0.0 | 0.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.5 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 5.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 7.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 2.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.7 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.4 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.1 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.0 | 0.1 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.0 | 0.6 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.9 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 1.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 3.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 4.8 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.5 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.9 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.3 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 5.5 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 1.7 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 1.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.3 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 1.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 1.8 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.4 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.3 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 1.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0004336 | galactosylceramidase activity(GO:0004336) |
| 1.3 | 3.8 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.8 | 11.8 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.7 | 2.8 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.6 | 3.9 | GO:0052845 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.6 | 3.1 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.6 | 1.7 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.5 | 1.6 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.5 | 1.4 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.5 | 2.7 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.4 | 2.6 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.4 | 1.9 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.4 | 1.2 | GO:0002135 | CTP binding(GO:0002135) |
| 0.4 | 1.4 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.4 | 1.8 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.3 | 3.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.3 | 2.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.3 | 1.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.3 | 2.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.3 | 0.9 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.3 | 1.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.3 | 6.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.3 | 2.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 4.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.3 | 1.6 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.3 | 5.6 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.3 | 9.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.3 | 1.0 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.2 | 1.2 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.2 | 0.9 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.2 | 2.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.2 | 0.7 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.2 | 0.9 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.2 | 0.7 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.2 | 1.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.2 | 3.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 0.7 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.2 | 5.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 1.5 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.2 | 1.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 4.3 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.2 | 2.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 1.0 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.2 | 0.6 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.2 | 7.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 1.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.2 | 1.3 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.2 | 3.8 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 0.5 | GO:0098973 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.2 | 2.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 2.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.2 | 0.9 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.2 | 2.9 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.2 | 0.5 | GO:0050693 | DBD domain binding(GO:0050692) LBD domain binding(GO:0050693) |
| 0.2 | 1.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.2 | 0.5 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.2 | 0.8 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 1.6 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 0.8 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.2 | 3.0 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.2 | 1.5 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.2 | 1.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 2.7 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.4 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.1 | 2.0 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 1.9 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.4 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.1 | 0.5 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 0.7 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.4 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.1 | 2.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.6 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.6 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.5 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 2.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.3 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 0.4 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.1 | 0.4 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.1 | 0.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 1.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.5 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 3.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 1.4 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.6 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.5 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 0.2 | GO:0033265 | choline binding(GO:0033265) |
| 0.1 | 0.6 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 5.7 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.1 | 0.4 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.1 | 1.7 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.4 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 0.9 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.1 | 0.6 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.1 | 0.8 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 1.3 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.1 | 0.3 | GO:0070404 | NADH binding(GO:0070404) |
| 0.1 | 0.3 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.1 | 0.6 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.1 | 0.8 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.1 | 0.5 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.1 | 1.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 0.3 | GO:0097604 | temperature-gated cation channel activity(GO:0097604) |
| 0.1 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 1.5 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 0.3 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 2.1 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 0.9 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.1 | 0.4 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 0.4 | GO:0004803 | transposase activity(GO:0004803) |
| 0.1 | 1.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 1.8 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 8.4 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 0.3 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 0.2 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.1 | 0.6 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 2.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 3.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.3 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.9 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.6 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 0.4 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.2 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.1 | 0.4 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 1.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.4 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.1 | 0.2 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.5 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.4 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 1.8 | GO:0035004 | phosphatidylinositol 3-kinase activity(GO:0035004) |
| 0.1 | 0.3 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 2.3 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.2 | GO:0005135 | erythropoietin receptor binding(GO:0005128) interleukin-3 receptor binding(GO:0005135) |
| 0.1 | 1.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.3 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 1.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.2 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.1 | 0.2 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 1.7 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 4.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.4 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 3.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.5 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 1.4 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.2 | GO:0019144 | ADP-sugar diphosphatase activity(GO:0019144) |
| 0.0 | 2.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 5.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.4 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.4 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.3 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.6 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.0 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.1 | GO:0016717 | C-5 sterol desaturase activity(GO:0000248) oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) sterol desaturase activity(GO:0070704) |
| 0.0 | 0.2 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 3.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 3.0 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.2 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.5 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.8 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.2 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 1.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.5 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 1.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.0 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 6.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.1 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 1.8 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 1.6 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 1.2 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 1.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.6 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.3 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 1.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.3 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.1 | GO:0046592 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.0 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.7 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 2.0 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 0.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.3 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.3 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.5 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.4 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 2.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 1.2 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.9 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 5.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.5 | GO:0019841 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.0 | 0.8 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.3 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 1.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 1.0 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.0 | 0.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.5 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.2 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.0 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.4 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 5.8 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 1.2 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 1.0 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.5 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.2 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.6 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.4 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.2 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0032396 | MHC class Ib protein binding(GO:0023029) inhibitory MHC class I receptor activity(GO:0032396) |
| 0.0 | 0.2 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 2.9 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 3.1 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.0 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.0 | 0.2 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 6.2 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.0 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 14.8 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.2 | 6.5 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.2 | 3.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 8.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 10.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 3.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 8.0 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 3.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 3.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 0.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 5.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 2.0 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 2.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 2.6 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 6.0 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 3.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 1.0 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 0.6 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 3.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 1.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 0.7 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 2.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 4.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 2.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 3.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 1.0 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 0.9 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 5.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 5.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 3.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 5.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 3.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.8 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 3.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.7 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 2.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.5 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.6 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.3 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.4 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.8 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.6 | 3.3 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.3 | 4.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.3 | 4.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 8.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.2 | 4.0 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 3.9 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.2 | 2.7 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 0.6 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.1 | 2.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.3 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.1 | 1.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 3.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 0.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 2.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 2.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 4.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 2.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.7 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 2.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 0.2 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.1 | 2.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 0.9 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 2.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 4.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 2.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.5 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 1.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.8 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 2.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 6.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.9 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.7 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.6 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 5.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.0 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 1.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 3.6 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.9 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.7 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 3.0 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.6 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.8 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 2.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 3.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.6 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.7 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.0 | 0.7 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 2.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.1 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 1.4 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.0 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 4.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.7 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 1.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.0 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.2 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 MUTANTS | Genes involved in Signaling by FGFR1 mutants |
| 0.0 | 0.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |