Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
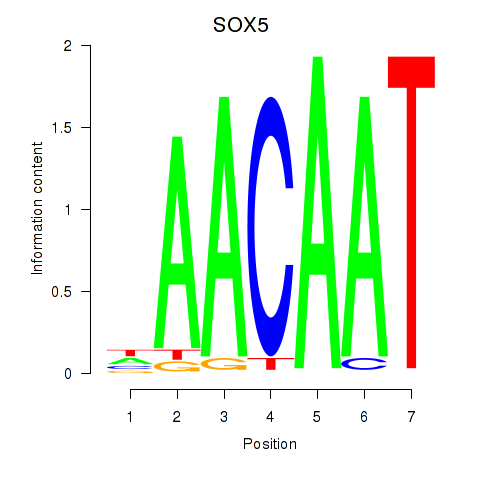
Results for SOX5
Z-value: 1.37
Transcription factors associated with SOX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX5
|
ENSG00000134532.19 | SOX5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX5 | hg38_v1_chr12_-_23951020_23951037 | 0.63 | 8.2e-04 | Click! |
Activity profile of SOX5 motif
Sorted Z-values of SOX5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_41612892 | 7.37 |
ENST00000509454.5
ENST00000396595.7 ENST00000381753.8 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr4_+_41612702 | 6.94 |
ENST00000509277.5
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr3_-_18424533 | 6.89 |
ENST00000417717.6
|
SATB1
|
SATB homeobox 1 |
| chr2_-_187448244 | 6.85 |
ENST00000392370.8
ENST00000410068.5 ENST00000447403.5 ENST00000410102.5 |
CALCRL
|
calcitonin receptor like receptor |
| chr7_-_11832190 | 6.41 |
ENST00000423059.9
ENST00000617773.1 |
THSD7A
|
thrombospondin type 1 domain containing 7A |
| chr4_-_100517991 | 5.57 |
ENST00000511970.5
ENST00000502569.1 ENST00000305864.7 ENST00000296420.9 |
EMCN
|
endomucin |
| chrX_-_117973579 | 5.18 |
ENST00000371878.5
|
KLHL13
|
kelch like family member 13 |
| chr18_+_44680875 | 4.94 |
ENST00000649279.2
ENST00000677699.1 |
SETBP1
|
SET binding protein 1 |
| chr4_-_185810894 | 4.63 |
ENST00000448662.6
ENST00000439049.5 ENST00000420158.5 ENST00000319471.13 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_+_61081728 | 4.38 |
ENST00000371189.8
|
NFIA
|
nuclear factor I A |
| chr10_-_114684612 | 4.16 |
ENST00000533213.6
ENST00000369252.8 |
ABLIM1
|
actin binding LIM protein 1 |
| chr21_+_17513003 | 3.97 |
ENST00000284878.12
ENST00000400166.5 |
CXADR
|
CXADR Ig-like cell adhesion molecule |
| chr9_-_14314519 | 3.97 |
ENST00000397581.6
|
NFIB
|
nuclear factor I B |
| chr2_-_156342348 | 3.96 |
ENST00000409572.5
|
NR4A2
|
nuclear receptor subfamily 4 group A member 2 |
| chr9_-_14314567 | 3.94 |
ENST00000397579.6
|
NFIB
|
nuclear factor I B |
| chr14_-_91947654 | 3.70 |
ENST00000342058.9
|
FBLN5
|
fibulin 5 |
| chr9_-_14314132 | 3.68 |
ENST00000380953.6
|
NFIB
|
nuclear factor I B |
| chr3_+_134795248 | 3.54 |
ENST00000398015.8
|
EPHB1
|
EPH receptor B1 |
| chr9_-_131270493 | 3.52 |
ENST00000372269.7
ENST00000464831.1 |
FAM78A
|
family with sequence similarity 78 member A |
| chr14_-_91946989 | 3.49 |
ENST00000556154.5
|
FBLN5
|
fibulin 5 |
| chr12_+_53050014 | 3.41 |
ENST00000314250.11
|
TNS2
|
tensin 2 |
| chrX_-_117973717 | 3.32 |
ENST00000262820.7
|
KLHL13
|
kelch like family member 13 |
| chr12_+_53050179 | 3.30 |
ENST00000546602.5
ENST00000552570.5 ENST00000549700.5 |
TNS2
|
tensin 2 |
| chr14_-_91947383 | 3.18 |
ENST00000267620.14
|
FBLN5
|
fibulin 5 |
| chr10_+_35126791 | 3.16 |
ENST00000474362.5
ENST00000374721.7 |
CREM
|
cAMP responsive element modulator |
| chr10_+_35127162 | 3.14 |
ENST00000354759.7
|
CREM
|
cAMP responsive element modulator |
| chr12_+_32502114 | 3.00 |
ENST00000682739.1
ENST00000427716.7 ENST00000583694.2 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr9_+_2159672 | 2.95 |
ENST00000634343.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr14_+_61187544 | 2.90 |
ENST00000555185.5
ENST00000557294.5 ENST00000556778.5 |
PRKCH
|
protein kinase C eta |
| chr1_+_185734362 | 2.84 |
ENST00000271588.9
|
HMCN1
|
hemicentin 1 |
| chr10_+_35127023 | 2.63 |
ENST00000429130.7
ENST00000469949.6 ENST00000460270.5 |
CREM
|
cAMP responsive element modulator |
| chr12_-_85836372 | 2.55 |
ENST00000361228.5
|
RASSF9
|
Ras association domain family member 9 |
| chr9_+_79573162 | 2.50 |
ENST00000425506.5
|
TLE4
|
TLE family member 4, transcriptional corepressor |
| chr1_+_164559739 | 2.47 |
ENST00000627490.2
|
PBX1
|
PBX homeobox 1 |
| chr15_-_52652031 | 2.41 |
ENST00000546305.6
|
FAM214A
|
family with sequence similarity 214 member A |
| chr14_-_89417148 | 2.32 |
ENST00000557258.6
|
FOXN3
|
forkhead box N3 |
| chr9_-_137302264 | 2.29 |
ENST00000356628.4
|
NRARP
|
NOTCH regulated ankyrin repeat protein |
| chr20_-_47356670 | 2.22 |
ENST00000540497.5
ENST00000461685.5 ENST00000617418.4 ENST00000435836.5 ENST00000471951.6 ENST00000352431.6 ENST00000360911.7 ENST00000458360.6 |
ZMYND8
|
zinc finger MYND-type containing 8 |
| chr2_+_69013170 | 2.19 |
ENST00000303714.9
|
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr3_-_142028597 | 2.14 |
ENST00000467667.5
|
TFDP2
|
transcription factor Dp-2 |
| chrX_+_136169624 | 2.13 |
ENST00000394153.6
|
FHL1
|
four and a half LIM domains 1 |
| chr2_-_159616442 | 2.12 |
ENST00000541068.6
ENST00000392783.7 ENST00000392782.5 |
BAZ2B
|
bromodomain adjacent to zinc finger domain 2B |
| chr5_-_88785493 | 2.07 |
ENST00000503554.4
|
MEF2C
|
myocyte enhancer factor 2C |
| chrX_+_136169664 | 2.02 |
ENST00000456445.5
|
FHL1
|
four and a half LIM domains 1 |
| chr10_+_35127295 | 1.95 |
ENST00000489321.5
ENST00000427847.6 ENST00000374728.7 ENST00000345491.7 ENST00000487132.5 |
CREM
|
cAMP responsive element modulator |
| chrX_+_136169833 | 1.94 |
ENST00000628032.2
|
FHL1
|
four and a half LIM domains 1 |
| chr9_-_122227525 | 1.94 |
ENST00000373755.6
ENST00000373754.6 |
LHX6
|
LIM homeobox 6 |
| chr20_-_47356721 | 1.93 |
ENST00000262975.8
ENST00000446994.6 ENST00000355972.8 ENST00000396281.8 ENST00000619049.4 ENST00000611941.4 ENST00000372023.7 |
ZMYND8
|
zinc finger MYND-type containing 8 |
| chr9_-_15510954 | 1.89 |
ENST00000380733.9
|
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr12_-_6124662 | 1.88 |
ENST00000261405.10
|
VWF
|
von Willebrand factor |
| chr3_+_134795277 | 1.82 |
ENST00000647596.1
|
EPHB1
|
EPH receptor B1 |
| chr9_-_123268538 | 1.81 |
ENST00000360998.3
ENST00000348403.10 |
STRBP
|
spermatid perinuclear RNA binding protein |
| chr18_-_55510753 | 1.79 |
ENST00000543082.5
|
TCF4
|
transcription factor 4 |
| chr7_-_32892015 | 1.79 |
ENST00000452926.1
|
KBTBD2
|
kelch repeat and BTB domain containing 2 |
| chr3_+_124033356 | 1.71 |
ENST00000682506.1
|
KALRN
|
kalirin RhoGEF kinase |
| chr17_+_69502397 | 1.67 |
ENST00000613873.4
ENST00000589647.5 |
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr2_-_110473041 | 1.65 |
ENST00000632897.1
ENST00000413601.3 |
LIMS4
|
LIM zinc finger domain containing 4 |
| chr8_+_67952028 | 1.61 |
ENST00000288368.5
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate dependent Rac exchange factor 2 |
| chr3_-_65597886 | 1.61 |
ENST00000460329.6
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr21_+_17513119 | 1.58 |
ENST00000356275.10
ENST00000400165.5 ENST00000400169.1 |
CXADR
|
CXADR Ig-like cell adhesion molecule |
| chr20_+_33562306 | 1.57 |
ENST00000344201.7
|
CBFA2T2
|
CBFA2/RUNX1 partner transcriptional co-repressor 2 |
| chrX_+_9912434 | 1.56 |
ENST00000418909.6
|
SHROOM2
|
shroom family member 2 |
| chr3_-_142028617 | 1.56 |
ENST00000477292.5
ENST00000478006.5 ENST00000495310.5 ENST00000486111.5 |
TFDP2
|
transcription factor Dp-2 |
| chr17_-_64390852 | 1.55 |
ENST00000563924.6
|
PECAM1
|
platelet and endothelial cell adhesion molecule 1 |
| chr3_-_71305986 | 1.53 |
ENST00000647614.1
|
FOXP1
|
forkhead box P1 |
| chr21_-_38498415 | 1.52 |
ENST00000398905.5
ENST00000398907.5 ENST00000453032.6 ENST00000288319.12 |
ERG
|
ETS transcription factor ERG |
| chr2_+_109898685 | 1.52 |
ENST00000480744.2
|
LIMS3
|
LIM zinc finger domain containing 3 |
| chr5_+_139341826 | 1.49 |
ENST00000265192.9
|
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr3_+_25428233 | 1.47 |
ENST00000437042.6
ENST00000330688.9 |
RARB
|
retinoic acid receptor beta |
| chr2_+_202634960 | 1.44 |
ENST00000392238.3
|
FAM117B
|
family with sequence similarity 117 member B |
| chr9_-_15510991 | 1.42 |
ENST00000380715.5
ENST00000380716.8 ENST00000380738.8 |
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr6_+_118894144 | 1.42 |
ENST00000229595.6
|
ASF1A
|
anti-silencing function 1A histone chaperone |
| chr20_-_47355657 | 1.42 |
ENST00000311275.11
|
ZMYND8
|
zinc finger MYND-type containing 8 |
| chr18_-_55321640 | 1.40 |
ENST00000637169.2
|
TCF4
|
transcription factor 4 |
| chr1_+_33256479 | 1.37 |
ENST00000539719.6
ENST00000483388.5 |
ZNF362
|
zinc finger protein 362 |
| chr4_+_169660062 | 1.32 |
ENST00000507875.5
ENST00000613795.4 |
CLCN3
|
chloride voltage-gated channel 3 |
| chr9_-_69672341 | 1.32 |
ENST00000265381.7
|
APBA1
|
amyloid beta precursor protein binding family A member 1 |
| chr5_+_139341875 | 1.32 |
ENST00000511706.5
|
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chrX_-_100636799 | 1.30 |
ENST00000373020.9
|
TSPAN6
|
tetraspanin 6 |
| chr12_-_70637405 | 1.30 |
ENST00000548122.2
ENST00000551525.5 ENST00000550358.5 ENST00000334414.11 |
PTPRB
|
protein tyrosine phosphatase receptor type B |
| chr20_+_325536 | 1.29 |
ENST00000342665.5
|
SOX12
|
SRY-box transcription factor 12 |
| chr14_-_21536884 | 1.28 |
ENST00000546363.5
|
SALL2
|
spalt like transcription factor 2 |
| chr7_+_77538027 | 1.24 |
ENST00000433369.6
ENST00000415482.6 |
PTPN12
|
protein tyrosine phosphatase non-receptor type 12 |
| chr6_+_155216637 | 1.23 |
ENST00000275246.11
|
TIAM2
|
TIAM Rac1 associated GEF 2 |
| chr18_-_55321986 | 1.21 |
ENST00000570287.6
|
TCF4
|
transcription factor 4 |
| chr9_+_27109393 | 1.18 |
ENST00000406359.8
|
TEK
|
TEK receptor tyrosine kinase |
| chr3_+_141386862 | 1.18 |
ENST00000513258.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr13_-_74133892 | 1.16 |
ENST00000377669.7
|
KLF12
|
Kruppel like factor 12 |
| chr12_+_59664677 | 1.15 |
ENST00000548610.5
|
SLC16A7
|
solute carrier family 16 member 7 |
| chr10_-_14572123 | 1.15 |
ENST00000378465.7
ENST00000452706.6 ENST00000622567.4 ENST00000378458.6 |
FAM107B
|
family with sequence similarity 107 member B |
| chr2_-_163735989 | 1.11 |
ENST00000333129.4
ENST00000409634.5 |
FIGN
|
fidgetin, microtubule severing factor |
| chr3_+_141387616 | 1.08 |
ENST00000509883.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr6_-_79234619 | 1.06 |
ENST00000344726.9
ENST00000275036.11 |
HMGN3
|
high mobility group nucleosomal binding domain 3 |
| chr9_-_95509241 | 1.05 |
ENST00000331920.11
|
PTCH1
|
patched 1 |
| chrX_+_108439779 | 1.05 |
ENST00000328300.11
|
COL4A5
|
collagen type IV alpha 5 chain |
| chr1_-_182391363 | 1.04 |
ENST00000417584.6
|
GLUL
|
glutamate-ammonia ligase |
| chr1_-_52552994 | 1.04 |
ENST00000355809.4
ENST00000528642.5 ENST00000470626.1 ENST00000257177.9 ENST00000371544.7 |
TUT4
|
terminal uridylyl transferase 4 |
| chr15_+_66702219 | 1.03 |
ENST00000288840.10
|
SMAD6
|
SMAD family member 6 |
| chrX_+_108439866 | 1.03 |
ENST00000361603.7
|
COL4A5
|
collagen type IV alpha 5 chain |
| chr10_+_61901678 | 1.03 |
ENST00000644638.1
ENST00000681100.1 ENST00000279873.12 |
ARID5B
|
AT-rich interaction domain 5B |
| chr2_-_70248598 | 1.01 |
ENST00000445587.5
ENST00000433529.7 ENST00000415783.6 |
TIA1
|
TIA1 cytotoxic granule associated RNA binding protein |
| chr18_-_55322215 | 1.00 |
ENST00000457482.7
|
TCF4
|
transcription factor 4 |
| chr6_-_79234713 | 1.00 |
ENST00000620514.1
|
HMGN3
|
high mobility group nucleosomal binding domain 3 |
| chr10_+_96043394 | 1.00 |
ENST00000403870.7
ENST00000265992.9 ENST00000465148.3 |
CCNJ
|
cyclin J |
| chr2_-_70248454 | 1.00 |
ENST00000416149.6
ENST00000282574.8 |
TIA1
|
TIA1 cytotoxic granule associated RNA binding protein |
| chr6_+_122399621 | 0.99 |
ENST00000368455.9
|
HSF2
|
heat shock transcription factor 2 |
| chr2_-_25982471 | 0.98 |
ENST00000264712.8
|
KIF3C
|
kinesin family member 3C |
| chr11_+_46381033 | 0.96 |
ENST00000359803.7
|
MDK
|
midkine |
| chr6_+_122399536 | 0.96 |
ENST00000452194.5
|
HSF2
|
heat shock transcription factor 2 |
| chr19_-_19515542 | 0.96 |
ENST00000585580.4
|
TSSK6
|
testis specific serine kinase 6 |
| chr6_+_36029082 | 0.94 |
ENST00000472333.1
|
MAPK14
|
mitogen-activated protein kinase 14 |
| chr12_+_55966821 | 0.94 |
ENST00000553376.5
ENST00000440311.6 ENST00000266970.9 ENST00000354056.4 |
CDK2
|
cyclin dependent kinase 2 |
| chr9_-_71768386 | 0.94 |
ENST00000377066.9
ENST00000377044.9 |
CEMIP2
|
cell migration inducing hyaluronidase 2 |
| chr5_+_139308095 | 0.93 |
ENST00000515833.2
|
MATR3
|
matrin 3 |
| chr9_-_120714457 | 0.92 |
ENST00000373930.4
|
MEGF9
|
multiple EGF like domains 9 |
| chrX_-_101617921 | 0.91 |
ENST00000361910.9
ENST00000538627.5 ENST00000539247.5 |
ARMCX6
|
armadillo repeat containing X-linked 6 |
| chr14_-_50561119 | 0.90 |
ENST00000555216.5
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr10_+_97319250 | 0.90 |
ENST00000371021.5
|
FRAT1
|
FRAT regulator of WNT signaling pathway 1 |
| chr3_-_69080350 | 0.88 |
ENST00000630585.1
ENST00000361055.9 ENST00000415609.6 ENST00000349511.8 |
UBA3
|
ubiquitin like modifier activating enzyme 3 |
| chr11_+_46380746 | 0.86 |
ENST00000405308.6
|
MDK
|
midkine |
| chr2_+_69013379 | 0.86 |
ENST00000409349.7
|
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr11_-_82997477 | 0.86 |
ENST00000534301.5
|
RAB30
|
RAB30, member RAS oncogene family |
| chr14_-_23435652 | 0.86 |
ENST00000355349.4
|
MYH7
|
myosin heavy chain 7 |
| chr5_-_38595396 | 0.85 |
ENST00000263409.8
|
LIFR
|
LIF receptor subunit alpha |
| chr3_-_48717166 | 0.83 |
ENST00000413654.5
ENST00000454335.5 ENST00000440424.5 ENST00000449610.5 ENST00000443964.1 ENST00000417896.1 ENST00000413298.5 ENST00000449563.5 ENST00000443853.5 ENST00000437427.5 ENST00000446860.5 ENST00000412850.5 ENST00000424035.1 ENST00000340879.8 ENST00000431721.6 ENST00000434860.1 ENST00000328631.10 ENST00000432678.6 |
IP6K2
|
inositol hexakisphosphate kinase 2 |
| chr11_-_102452758 | 0.83 |
ENST00000398136.7
ENST00000361236.7 |
TMEM123
|
transmembrane protein 123 |
| chr3_-_114624193 | 0.82 |
ENST00000481632.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chrX_-_112840815 | 0.82 |
ENST00000304758.5
ENST00000371959.9 |
AMOT
|
angiomotin |
| chr11_+_46381194 | 0.81 |
ENST00000533952.5
|
MDK
|
midkine |
| chr9_-_70869076 | 0.80 |
ENST00000677594.1
|
TRPM3
|
transient receptor potential cation channel subfamily M member 3 |
| chr1_+_81306096 | 0.76 |
ENST00000370721.5
ENST00000370727.5 ENST00000370725.5 ENST00000370723.5 ENST00000370728.5 ENST00000370730.5 |
ADGRL2
|
adhesion G protein-coupled receptor L2 |
| chr1_+_150364136 | 0.75 |
ENST00000369068.5
|
RPRD2
|
regulation of nuclear pre-mRNA domain containing 2 |
| chr15_+_43792839 | 0.75 |
ENST00000409614.1
|
SERF2
|
small EDRK-rich factor 2 |
| chr1_-_150268941 | 0.74 |
ENST00000369109.8
ENST00000236017.5 |
APH1A
|
aph-1 homolog A, gamma-secretase subunit |
| chr12_-_102480604 | 0.74 |
ENST00000392905.7
|
IGF1
|
insulin like growth factor 1 |
| chr11_+_46380932 | 0.73 |
ENST00000441869.5
|
MDK
|
midkine |
| chr18_+_58864866 | 0.72 |
ENST00000588456.5
ENST00000591808.6 ENST00000589481.1 ENST00000591049.1 |
ZNF532
|
zinc finger protein 532 |
| chr5_+_139342442 | 0.69 |
ENST00000394795.6
ENST00000510080.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chrX_-_50814095 | 0.69 |
ENST00000376020.8
|
SHROOM4
|
shroom family member 4 |
| chr12_-_102197827 | 0.68 |
ENST00000329406.5
|
PMCH
|
pro-melanin concentrating hormone |
| chr1_+_206834347 | 0.68 |
ENST00000340758.7
|
IL19
|
interleukin 19 |
| chr2_-_197675578 | 0.68 |
ENST00000295049.9
|
RFTN2
|
raftlin family member 2 |
| chrX_-_64976500 | 0.68 |
ENST00000447788.6
|
ZC4H2
|
zinc finger C4H2-type containing |
| chr12_-_31590967 | 0.67 |
ENST00000354285.8
|
DENND5B
|
DENN domain containing 5B |
| chr1_-_85578345 | 0.66 |
ENST00000426972.8
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr3_+_189789672 | 0.65 |
ENST00000434928.5
|
TP63
|
tumor protein p63 |
| chr16_-_1782526 | 0.64 |
ENST00000566339.6
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chr10_+_21524670 | 0.64 |
ENST00000631589.1
|
MLLT10
|
MLLT10 histone lysine methyltransferase DOT1L cofactor |
| chrX_-_24672654 | 0.63 |
ENST00000379145.5
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr1_+_150364621 | 0.63 |
ENST00000401000.8
|
RPRD2
|
regulation of nuclear pre-mRNA domain containing 2 |
| chr6_+_43059857 | 0.63 |
ENST00000259708.7
ENST00000472792.1 ENST00000479388.5 ENST00000460283.1 ENST00000394056.6 |
KLC4
|
kinesin light chain 4 |
| chr2_+_33436304 | 0.62 |
ENST00000402538.7
|
RASGRP3
|
RAS guanyl releasing protein 3 |
| chr7_-_27165517 | 0.62 |
ENST00000396345.1
ENST00000343483.7 |
HOXA9
|
homeobox A9 |
| chr6_+_89080739 | 0.62 |
ENST00000369472.1
ENST00000336032.4 |
PNRC1
|
proline rich nuclear receptor coactivator 1 |
| chr16_+_1078781 | 0.61 |
ENST00000293897.5
|
SSTR5
|
somatostatin receptor 5 |
| chr6_-_90296908 | 0.61 |
ENST00000537989.5
|
BACH2
|
BTB domain and CNC homolog 2 |
| chrX_+_123960519 | 0.60 |
ENST00000455404.5
ENST00000218089.13 |
STAG2
|
stromal antigen 2 |
| chr2_+_86720282 | 0.60 |
ENST00000283632.5
|
RMND5A
|
required for meiotic nuclear division 5 homolog A |
| chr17_+_39738317 | 0.59 |
ENST00000394211.7
|
GRB7
|
growth factor receptor bound protein 7 |
| chr11_+_128694052 | 0.58 |
ENST00000527786.7
ENST00000534087.3 |
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr6_-_99425269 | 0.57 |
ENST00000647811.1
ENST00000481229.2 ENST00000369239.10 ENST00000681611.1 ENST00000681615.1 ENST00000438806.5 |
PNISR
|
PNN interacting serine and arginine rich protein |
| chr10_+_21524627 | 0.57 |
ENST00000651097.1
|
MLLT10
|
MLLT10 histone lysine methyltransferase DOT1L cofactor |
| chr2_-_199458689 | 0.57 |
ENST00000443023.5
|
SATB2
|
SATB homeobox 2 |
| chr1_+_202193791 | 0.55 |
ENST00000367278.8
|
LGR6
|
leucine rich repeat containing G protein-coupled receptor 6 |
| chr3_+_14947680 | 0.55 |
ENST00000435454.5
ENST00000323373.10 |
NR2C2
|
nuclear receptor subfamily 2 group C member 2 |
| chr11_+_128693887 | 0.54 |
ENST00000281428.12
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr3_+_14947568 | 0.53 |
ENST00000413118.5
ENST00000425241.5 |
NR2C2
|
nuclear receptor subfamily 2 group C member 2 |
| chr12_-_48570046 | 0.53 |
ENST00000301046.6
ENST00000549817.1 |
LALBA
|
lactalbumin alpha |
| chr6_+_50818701 | 0.53 |
ENST00000344788.7
|
TFAP2B
|
transcription factor AP-2 beta |
| chr1_+_200739542 | 0.53 |
ENST00000358823.6
|
CAMSAP2
|
calmodulin regulated spectrin associated protein family member 2 |
| chr6_+_156777366 | 0.52 |
ENST00000636930.2
|
ARID1B
|
AT-rich interaction domain 1B |
| chr15_-_34318761 | 0.52 |
ENST00000290209.9
|
SLC12A6
|
solute carrier family 12 member 6 |
| chr22_-_46537593 | 0.51 |
ENST00000262738.9
ENST00000674500.2 |
CELSR1
|
cadherin EGF LAG seven-pass G-type receptor 1 |
| chrX_+_71283186 | 0.51 |
ENST00000535149.5
|
NONO
|
non-POU domain containing octamer binding |
| chrX_-_50814302 | 0.51 |
ENST00000289292.11
|
SHROOM4
|
shroom family member 4 |
| chr1_+_226063466 | 0.50 |
ENST00000666609.1
ENST00000661429.1 |
H3-3A
|
H3.3 histone A |
| chr4_+_109827963 | 0.50 |
ENST00000317735.7
|
RRH
|
retinal pigment epithelium-derived rhodopsin homolog |
| chrX_-_107716401 | 0.50 |
ENST00000486554.1
ENST00000372390.8 |
TSC22D3
|
TSC22 domain family member 3 |
| chr6_+_156777882 | 0.50 |
ENST00000350026.10
ENST00000647938.1 ENST00000674298.1 |
ARID1B
|
AT-rich interaction domain 1B |
| chr12_-_8662881 | 0.49 |
ENST00000433590.6
|
MFAP5
|
microfibril associated protein 5 |
| chr7_-_32891744 | 0.49 |
ENST00000304056.9
|
KBTBD2
|
kelch repeat and BTB domain containing 2 |
| chrX_-_17860707 | 0.49 |
ENST00000360011.5
|
RAI2
|
retinoic acid induced 2 |
| chr1_+_28914614 | 0.47 |
ENST00000645184.1
|
EPB41
|
erythrocyte membrane protein band 4.1 |
| chr8_+_102551583 | 0.46 |
ENST00000285402.4
|
ODF1
|
outer dense fiber of sperm tails 1 |
| chr9_+_27109135 | 0.46 |
ENST00000519097.5
ENST00000615002.4 |
TEK
|
TEK receptor tyrosine kinase |
| chr3_+_19947316 | 0.46 |
ENST00000422242.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr15_-_37099306 | 0.45 |
ENST00000557796.6
ENST00000397620.6 |
MEIS2
|
Meis homeobox 2 |
| chrX_+_21940693 | 0.45 |
ENST00000404933.7
ENST00000379404.5 |
SMS
|
spermine synthase |
| chr1_+_228149922 | 0.44 |
ENST00000366714.3
|
GJC2
|
gap junction protein gamma 2 |
| chr9_+_27109200 | 0.44 |
ENST00000380036.10
|
TEK
|
TEK receptor tyrosine kinase |
| chr1_-_224433776 | 0.43 |
ENST00000678879.1
ENST00000651911.2 |
WDR26
|
WD repeat domain 26 |
| chr1_-_150269051 | 0.42 |
ENST00000414276.6
ENST00000360244.8 |
APH1A
|
aph-1 homolog A, gamma-secretase subunit |
| chr1_-_151146643 | 0.42 |
ENST00000613223.1
|
SEMA6C
|
semaphorin 6C |
| chr15_+_98648502 | 0.41 |
ENST00000650285.1
ENST00000649865.1 |
IGF1R
|
insulin like growth factor 1 receptor |
| chr10_-_11611754 | 0.41 |
ENST00000609104.5
|
USP6NL
|
USP6 N-terminal like |
| chr3_-_115071333 | 0.41 |
ENST00000462705.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr17_-_45432916 | 0.40 |
ENST00000290470.3
|
ARHGAP27
|
Rho GTPase activating protein 27 |
| chr20_-_62427528 | 0.40 |
ENST00000252998.2
|
RBBP8NL
|
RBBP8 N-terminal like |
| chr18_+_3451647 | 0.40 |
ENST00000345133.9
ENST00000330513.10 ENST00000549546.5 |
TGIF1
|
TGFB induced factor homeobox 1 |
| chr1_-_156248013 | 0.38 |
ENST00000368270.2
|
PAQR6
|
progestin and adipoQ receptor family member 6 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:0099557 | trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission(GO:0099557) |
| 1.4 | 5.6 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 1.3 | 4.0 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 1.1 | 5.5 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 1.1 | 11.6 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.9 | 10.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.6 | 3.0 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.6 | 1.7 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.5 | 6.9 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.5 | 3.4 | GO:0030421 | defecation(GO:0030421) |
| 0.4 | 6.9 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.4 | 1.6 | GO:0072011 | diapedesis(GO:0050904) glomerular endothelium development(GO:0072011) |
| 0.4 | 3.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.3 | 2.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.3 | 1.3 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.3 | 1.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.3 | 2.9 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.3 | 1.0 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.3 | 2.1 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.3 | 2.3 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.3 | 1.5 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.2 | 0.9 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.2 | 0.9 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.2 | 0.4 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.2 | 6.7 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.2 | 0.6 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.2 | 0.5 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.2 | 0.5 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.2 | 1.7 | GO:0046959 | habituation(GO:0046959) negative regulation of growth hormone secretion(GO:0060125) |
| 0.2 | 12.2 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.2 | 0.9 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.2 | 0.9 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.2 | 0.8 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.1 | 1.9 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 2.1 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.1 | 1.1 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.8 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 3.8 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 2.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 0.4 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 4.3 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 1.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 1.0 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.1 | 0.6 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.1 | 2.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.8 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 1.0 | GO:0003183 | mitral valve development(GO:0003174) mitral valve morphogenesis(GO:0003183) |
| 0.1 | 0.3 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.1 | 0.3 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 1.3 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 3.5 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 2.4 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 0.6 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.3 | GO:0097274 | ammonia homeostasis(GO:0097272) urea homeostasis(GO:0097274) |
| 0.1 | 0.3 | GO:0030047 | actin modification(GO:0030047) |
| 0.1 | 1.0 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 | 2.5 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.1 | 3.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 1.3 | GO:1901223 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.4 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.7 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 0.5 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 1.9 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 1.0 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.1 | 1.5 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.2 | GO:0021658 | rhombomere morphogenesis(GO:0021593) rhombomere 3 morphogenesis(GO:0021658) |
| 0.1 | 6.0 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.1 | 0.3 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.1 | 0.6 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.7 | GO:0032596 | protein transport into membrane raft(GO:0032596) dsRNA transport(GO:0033227) |
| 0.1 | 1.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.8 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.6 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.4 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.1 | 1.1 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.5 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.8 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 1.1 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.6 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.7 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 0.3 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.1 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.0 | 0.7 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.0 | 0.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 12.2 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 1.1 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.7 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 | 0.6 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.0 | 0.1 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 6.1 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 0.1 | GO:0021834 | embryonic olfactory bulb interneuron precursor migration(GO:0021831) chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 1.6 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.1 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 1.2 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 6.7 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.7 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 1.0 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 2.1 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.2 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.0 | 0.0 | GO:0038031 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 4.0 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.2 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 2.5 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.4 | GO:0035886 | vascular smooth muscle cell differentiation(GO:0035886) |
| 0.0 | 3.3 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.3 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 1.0 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 1.7 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.7 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 0.1 | GO:0002399 | MHC protein complex assembly(GO:0002396) MHC class II protein complex assembly(GO:0002399) |
| 0.0 | 0.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 1.0 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.0 | 0.8 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.4 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.7 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.1 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.7 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 5.2 | GO:0001525 | angiogenesis(GO:0001525) |
| 0.0 | 0.4 | GO:0010644 | cell communication by electrical coupling(GO:0010644) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 10.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.4 | 11.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.3 | 0.9 | GO:0097135 | X chromosome(GO:0000805) cyclin E2-CDK2 complex(GO:0097135) |
| 0.3 | 2.0 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.2 | 2.0 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.2 | 0.6 | GO:0034657 | GID complex(GO:0034657) |
| 0.2 | 5.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 6.6 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 1.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 2.6 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 11.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 3.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 1.6 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.4 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 0.7 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.1 | 0.8 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 2.9 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 3.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 4.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 1.0 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 1.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 6.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 18.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 1.1 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.7 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.5 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 1.1 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 3.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.4 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 1.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 14.5 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 1.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 2.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 6.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 2.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.3 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.9 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.6 | 10.8 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.6 | 5.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.5 | 5.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.5 | 2.9 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.4 | 1.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.3 | 1.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.3 | 1.0 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.3 | 5.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.3 | 3.3 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.2 | 0.8 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.2 | 1.1 | GO:0005119 | smoothened binding(GO:0005119) hedgehog family protein binding(GO:0097108) |
| 0.2 | 0.8 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.2 | 2.0 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.2 | 0.8 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.2 | 5.4 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.2 | 4.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.6 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.4 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 5.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.5 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 0.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.6 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 2.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 1.0 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.1 | 0.9 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 1.6 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.8 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.4 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.1 | 3.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.7 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 3.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 2.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 13.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 4.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 0.5 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.9 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 2.1 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 2.9 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 2.1 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.0 | 0.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.9 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 19.2 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 1.0 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 1.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.5 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 4.2 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.7 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.3 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 3.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 1.4 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 2.2 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 2.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.8 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.0 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 7.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.8 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 2.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 5.1 | GO:0003682 | chromatin binding(GO:0003682) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 16.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.2 | 10.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 4.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 4.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 4.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 1.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 2.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 0.7 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 3.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.7 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 4.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 6.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 3.1 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 1.6 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 3.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 2.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.2 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 9.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 3.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 5.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 2.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.8 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.8 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.9 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 3.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 0.9 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.3 | 11.6 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.3 | 3.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 1.6 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 1.9 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 4.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 8.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.7 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.1 | 7.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 5.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 2.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 7.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 5.5 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 0.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.5 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 4.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 2.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 8.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 2.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |