Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
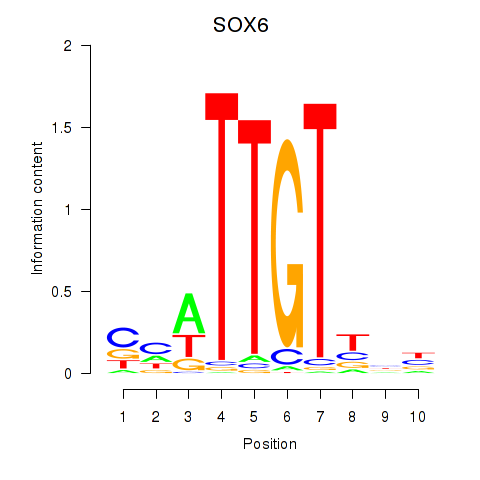
Results for SOX6
Z-value: 0.62
Transcription factors associated with SOX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX6
|
ENSG00000110693.19 | SOX6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX6 | hg38_v1_chr11_-_16408853_16408894 | 0.09 | 6.7e-01 | Click! |
Activity profile of SOX6 motif
Sorted Z-values of SOX6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_41612892 | 3.06 |
ENST00000509454.5
ENST00000396595.7 ENST00000381753.8 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr4_+_41612702 | 3.02 |
ENST00000509277.5
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr1_+_99646025 | 2.87 |
ENST00000263174.9
ENST00000605497.5 ENST00000615664.1 |
PALMD
|
palmdelphin |
| chr3_-_18424533 | 2.03 |
ENST00000417717.6
|
SATB1
|
SATB homeobox 1 |
| chr4_-_185810894 | 1.78 |
ENST00000448662.6
ENST00000439049.5 ENST00000420158.5 ENST00000319471.13 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr16_-_46831043 | 1.24 |
ENST00000565112.1
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr7_+_116672187 | 1.15 |
ENST00000318493.11
ENST00000397752.8 |
MET
|
MET proto-oncogene, receptor tyrosine kinase |
| chr20_-_40689228 | 1.14 |
ENST00000373313.3
|
MAFB
|
MAF bZIP transcription factor B |
| chr18_+_6729698 | 1.13 |
ENST00000383472.9
|
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr9_-_137302264 | 1.10 |
ENST00000356628.4
|
NRARP
|
NOTCH regulated ankyrin repeat protein |
| chr4_-_185956348 | 1.09 |
ENST00000431902.5
ENST00000284776.11 ENST00000415274.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr22_+_19760714 | 1.02 |
ENST00000649276.2
|
TBX1
|
T-box transcription factor 1 |
| chrX_-_117973579 | 1.02 |
ENST00000371878.5
|
KLHL13
|
kelch like family member 13 |
| chr14_-_50561119 | 0.96 |
ENST00000555216.5
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr2_+_69013170 | 0.89 |
ENST00000303714.9
|
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr16_+_30395400 | 0.77 |
ENST00000320159.2
ENST00000613509.2 |
ZNF48
|
zinc finger protein 48 |
| chr4_+_54657918 | 0.76 |
ENST00000412167.6
ENST00000288135.6 |
KIT
|
KIT proto-oncogene, receptor tyrosine kinase |
| chr1_+_228149922 | 0.72 |
ENST00000366714.3
|
GJC2
|
gap junction protein gamma 2 |
| chr1_-_25906457 | 0.70 |
ENST00000426559.6
|
STMN1
|
stathmin 1 |
| chr17_-_64390852 | 0.68 |
ENST00000563924.6
|
PECAM1
|
platelet and endothelial cell adhesion molecule 1 |
| chr4_+_143381939 | 0.65 |
ENST00000505913.5
|
GAB1
|
GRB2 associated binding protein 1 |
| chr13_-_100674774 | 0.61 |
ENST00000328767.9
|
TMTC4
|
transmembrane O-mannosyltransferase targeting cadherins 4 |
| chr14_-_87993143 | 0.59 |
ENST00000622264.4
|
GALC
|
galactosylceramidase |
| chr14_+_103123452 | 0.58 |
ENST00000558056.1
ENST00000560869.6 |
TNFAIP2
|
TNF alpha induced protein 2 |
| chr16_+_29455105 | 0.57 |
ENST00000567248.1
|
SLX1B
|
SLX1 homolog B, structure-specific endonuclease subunit |
| chr7_+_77538027 | 0.57 |
ENST00000433369.6
ENST00000415482.6 |
PTPN12
|
protein tyrosine phosphatase non-receptor type 12 |
| chr17_+_69502397 | 0.56 |
ENST00000613873.4
ENST00000589647.5 |
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr9_-_131270493 | 0.56 |
ENST00000372269.7
ENST00000464831.1 |
FAM78A
|
family with sequence similarity 78 member A |
| chr2_-_207167220 | 0.51 |
ENST00000421199.5
ENST00000457962.5 |
KLF7
|
Kruppel like factor 7 |
| chr2_-_207166818 | 0.51 |
ENST00000423015.5
|
KLF7
|
Kruppel like factor 7 |
| chr8_-_28386417 | 0.51 |
ENST00000521185.5
ENST00000520290.5 ENST00000344423.10 |
ZNF395
|
zinc finger protein 395 |
| chr13_-_100674787 | 0.51 |
ENST00000342624.10
|
TMTC4
|
transmembrane O-mannosyltransferase targeting cadherins 4 |
| chr8_-_6563044 | 0.50 |
ENST00000338312.10
|
ANGPT2
|
angiopoietin 2 |
| chrX_-_45200895 | 0.50 |
ENST00000377934.4
|
DIPK2B
|
divergent protein kinase domain 2B |
| chr12_-_108826161 | 0.50 |
ENST00000546697.1
|
SSH1
|
slingshot protein phosphatase 1 |
| chr7_+_102912983 | 0.50 |
ENST00000339431.9
ENST00000249377.4 |
LRRC17
|
leucine rich repeat containing 17 |
| chr5_-_39425187 | 0.49 |
ENST00000545653.5
|
DAB2
|
DAB adaptor protein 2 |
| chr5_+_141359970 | 0.49 |
ENST00000522605.2
ENST00000622527.1 |
PCDHGB2
|
protocadherin gamma subfamily B, 2 |
| chr5_-_39424966 | 0.47 |
ENST00000515700.5
ENST00000320816.11 ENST00000339788.10 |
DAB2
|
DAB adaptor protein 2 |
| chr5_-_88785493 | 0.47 |
ENST00000503554.4
|
MEF2C
|
myocyte enhancer factor 2C |
| chr1_+_182839338 | 0.47 |
ENST00000367549.4
|
DHX9
|
DExH-box helicase 9 |
| chrX_-_45200828 | 0.47 |
ENST00000398000.7
|
DIPK2B
|
divergent protein kinase domain 2B |
| chr2_-_151828408 | 0.45 |
ENST00000295087.13
|
ARL5A
|
ADP ribosylation factor like GTPase 5A |
| chr5_+_141223332 | 0.45 |
ENST00000239449.7
ENST00000624896.1 ENST00000624396.1 |
PCDHB14
ENSG00000279983.1
|
protocadherin beta 14 novel protein |
| chr15_+_76931704 | 0.44 |
ENST00000320963.9
ENST00000394885.8 ENST00000394883.3 |
RCN2
|
reticulocalbin 2 |
| chr21_+_6111123 | 0.44 |
ENST00000613488.3
|
SIK1B
|
salt inducible kinase 1B (putative) |
| chr16_+_3018390 | 0.43 |
ENST00000573001.5
|
TNFRSF12A
|
TNF receptor superfamily member 12A |
| chr8_-_28386073 | 0.43 |
ENST00000523095.5
ENST00000522795.1 |
ZNF395
|
zinc finger protein 395 |
| chr14_-_87992838 | 0.42 |
ENST00000544807.6
|
GALC
|
galactosylceramidase |
| chr20_+_6767678 | 0.42 |
ENST00000378827.5
|
BMP2
|
bone morphogenetic protein 2 |
| chr14_-_87993159 | 0.42 |
ENST00000393568.8
ENST00000261304.7 |
GALC
|
galactosylceramidase |
| chr2_-_86563470 | 0.41 |
ENST00000409225.2
|
CHMP3
|
charged multivesicular body protein 3 |
| chr18_-_55321640 | 0.41 |
ENST00000637169.2
|
TCF4
|
transcription factor 4 |
| chr8_-_6563238 | 0.41 |
ENST00000629816.3
ENST00000523120.2 |
ANGPT2
|
angiopoietin 2 |
| chr11_+_128694052 | 0.40 |
ENST00000527786.7
ENST00000534087.3 |
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr6_-_75206044 | 0.40 |
ENST00000322507.13
|
COL12A1
|
collagen type XII alpha 1 chain |
| chr4_+_70704713 | 0.40 |
ENST00000417478.6
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr9_-_120714457 | 0.39 |
ENST00000373930.4
|
MEGF9
|
multiple EGF like domains 9 |
| chr8_-_6563409 | 0.39 |
ENST00000325203.9
|
ANGPT2
|
angiopoietin 2 |
| chr1_+_64470120 | 0.38 |
ENST00000651257.2
|
CACHD1
|
cache domain containing 1 |
| chr19_+_5681000 | 0.38 |
ENST00000581893.5
ENST00000411793.6 ENST00000301382.8 ENST00000581773.5 ENST00000339423.7 ENST00000423665.6 ENST00000583928.5 ENST00000342970.6 ENST00000422535.6 ENST00000581521.5 |
HSD11B1L
|
hydroxysteroid 11-beta dehydrogenase 1 like |
| chr19_-_17264718 | 0.37 |
ENST00000431146.6
ENST00000594190.5 |
USHBP1
|
USH1 protein network component harmonin binding protein 1 |
| chr16_-_15643024 | 0.37 |
ENST00000540441.6
|
MARF1
|
meiosis regulator and mRNA stability factor 1 |
| chr4_+_53377749 | 0.37 |
ENST00000507166.5
|
ENSG00000282278.1
|
novel FIP1L1-PDGFRA fusion protein |
| chr11_+_128693887 | 0.37 |
ENST00000281428.12
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr7_+_16753731 | 0.37 |
ENST00000262067.5
|
TSPAN13
|
tetraspanin 13 |
| chr10_-_13348270 | 0.37 |
ENST00000378614.8
ENST00000327347.10 ENST00000545675.5 |
SEPHS1
|
selenophosphate synthetase 1 |
| chrX_+_85244075 | 0.36 |
ENST00000276123.7
|
ZNF711
|
zinc finger protein 711 |
| chr11_-_10808304 | 0.36 |
ENST00000532082.6
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma 2 |
| chr3_-_52685794 | 0.36 |
ENST00000424867.1
ENST00000394830.7 ENST00000431678.5 ENST00000450271.5 |
PBRM1
|
polybromo 1 |
| chr10_-_24952573 | 0.36 |
ENST00000376378.5
ENST00000376376.3 ENST00000320152.11 |
PRTFDC1
|
phosphoribosyl transferase domain containing 1 |
| chr19_+_10602436 | 0.35 |
ENST00000590382.5
ENST00000407327.8 |
SLC44A2
|
solute carrier family 44 member 2 |
| chr15_+_81000913 | 0.35 |
ENST00000267984.4
|
TLNRD1
|
talin rod domain containing 1 |
| chr14_+_22281097 | 0.35 |
ENST00000390465.2
|
TRAV38-2DV8
|
T cell receptor alpha variable 38-2/delta variable 8 |
| chr1_-_201154418 | 0.34 |
ENST00000435310.5
ENST00000485839.6 |
TMEM9
|
transmembrane protein 9 |
| chr9_-_71768386 | 0.34 |
ENST00000377066.9
ENST00000377044.9 |
CEMIP2
|
cell migration inducing hyaluronidase 2 |
| chrX_+_85243983 | 0.34 |
ENST00000674551.1
|
ZNF711
|
zinc finger protein 711 |
| chr3_-_65597886 | 0.33 |
ENST00000460329.6
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr11_+_5488685 | 0.33 |
ENST00000322641.5
|
OR52D1
|
olfactory receptor family 52 subfamily D member 1 |
| chr3_+_25428233 | 0.32 |
ENST00000437042.6
ENST00000330688.9 |
RARB
|
retinoic acid receptor beta |
| chr9_-_70869076 | 0.32 |
ENST00000677594.1
|
TRPM3
|
transient receptor potential cation channel subfamily M member 3 |
| chr9_-_72364504 | 0.31 |
ENST00000237937.7
ENST00000343431.6 ENST00000376956.3 |
ZFAND5
|
zinc finger AN1-type containing 5 |
| chrX_+_85244032 | 0.31 |
ENST00000373165.7
|
ZNF711
|
zinc finger protein 711 |
| chr1_-_201154459 | 0.30 |
ENST00000414605.2
ENST00000367330.6 ENST00000367334.9 ENST00000367332.5 |
TMEM9
|
transmembrane protein 9 |
| chr7_-_99408548 | 0.29 |
ENST00000626285.1
ENST00000350498.8 |
PDAP1
|
PDGFA associated protein 1 |
| chr9_-_122227525 | 0.29 |
ENST00000373755.6
ENST00000373754.6 |
LHX6
|
LIM homeobox 6 |
| chr2_+_10123171 | 0.29 |
ENST00000615152.5
|
RRM2
|
ribonucleotide reductase regulatory subunit M2 |
| chr1_+_13585453 | 0.28 |
ENST00000487038.5
ENST00000475043.5 |
PDPN
|
podoplanin |
| chr3_+_183635605 | 0.28 |
ENST00000493074.1
ENST00000437402.5 ENST00000454495.6 ENST00000473045.5 ENST00000468101.5 ENST00000427201.2 ENST00000482138.1 ENST00000454652.6 ENST00000242810.11 |
KLHL24
|
kelch like family member 24 |
| chr19_+_46602050 | 0.28 |
ENST00000599839.5
ENST00000596362.1 |
CALM3
|
calmodulin 3 |
| chr11_-_82997394 | 0.27 |
ENST00000525117.5
ENST00000532548.5 |
RAB30
|
RAB30, member RAS oncogene family |
| chr11_-_82997477 | 0.27 |
ENST00000534301.5
|
RAB30
|
RAB30, member RAS oncogene family |
| chr18_-_55321986 | 0.27 |
ENST00000570287.6
|
TCF4
|
transcription factor 4 |
| chr8_-_94896660 | 0.27 |
ENST00000520509.5
|
CCNE2
|
cyclin E2 |
| chr6_+_125919296 | 0.27 |
ENST00000444128.2
|
NCOA7
|
nuclear receptor coactivator 7 |
| chrX_+_81202066 | 0.27 |
ENST00000373212.6
|
SH3BGRL
|
SH3 domain binding glutamate rich protein like |
| chr19_-_17264732 | 0.27 |
ENST00000252597.8
|
USHBP1
|
USH1 protein network component harmonin binding protein 1 |
| chr18_+_13218769 | 0.26 |
ENST00000677055.1
ENST00000399848.7 |
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr12_+_99647749 | 0.26 |
ENST00000324341.2
|
FAM71C
|
family with sequence similarity 71 member C |
| chrX_+_71283577 | 0.26 |
ENST00000420903.6
ENST00000373856.8 ENST00000678437.1 ENST00000678830.1 ENST00000677879.1 ENST00000373841.5 ENST00000276079.13 ENST00000676797.1 ENST00000678660.1 ENST00000678231.1 ENST00000677612.1 ENST00000413858.5 ENST00000450092.6 |
NONO
|
non-POU domain containing octamer binding |
| chr6_+_125919210 | 0.26 |
ENST00000438495.6
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr16_+_3654683 | 0.26 |
ENST00000246949.10
|
DNASE1
|
deoxyribonuclease 1 |
| chr11_-_44950151 | 0.26 |
ENST00000533940.5
ENST00000533937.1 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr14_-_74875998 | 0.25 |
ENST00000556489.4
ENST00000673765.1 |
PROX2
|
prospero homeobox 2 |
| chr5_-_180591488 | 0.25 |
ENST00000292641.4
|
SCGB3A1
|
secretoglobin family 3A member 1 |
| chr15_-_72783611 | 0.25 |
ENST00000563907.5
|
ADPGK
|
ADP dependent glucokinase |
| chr7_-_128775793 | 0.25 |
ENST00000249389.3
|
OPN1SW
|
opsin 1, short wave sensitive |
| chr13_+_111115303 | 0.25 |
ENST00000646102.2
ENST00000449979.5 |
ARHGEF7
|
Rho guanine nucleotide exchange factor 7 |
| chr6_+_32969345 | 0.24 |
ENST00000678250.1
|
BRD2
|
bromodomain containing 2 |
| chr4_-_22443110 | 0.24 |
ENST00000508133.5
|
ADGRA3
|
adhesion G protein-coupled receptor A3 |
| chr8_-_140800535 | 0.24 |
ENST00000521986.5
ENST00000523539.5 |
PTK2
|
protein tyrosine kinase 2 |
| chr7_+_107583919 | 0.24 |
ENST00000491150.5
|
BCAP29
|
B cell receptor associated protein 29 |
| chr4_-_137532452 | 0.24 |
ENST00000412923.6
ENST00000511115.5 ENST00000344876.9 ENST00000507846.5 ENST00000510305.5 ENST00000611581.1 |
PCDH18
|
protocadherin 18 |
| chr4_+_26584064 | 0.24 |
ENST00000264866.9
ENST00000505206.5 ENST00000511789.5 |
TBC1D19
|
TBC1 domain family member 19 |
| chr5_+_141172637 | 0.24 |
ENST00000231137.6
|
PCDHB7
|
protocadherin beta 7 |
| chr1_-_156859087 | 0.24 |
ENST00000368195.4
|
INSRR
|
insulin receptor related receptor |
| chr9_+_88388356 | 0.24 |
ENST00000375859.4
|
SPIN1
|
spindlin 1 |
| chr2_-_75560893 | 0.23 |
ENST00000410113.5
ENST00000393913.8 |
EVA1A
|
eva-1 homolog A, regulator of programmed cell death |
| chr1_+_15758768 | 0.23 |
ENST00000483633.6
ENST00000502739.5 ENST00000375766.8 ENST00000431771.6 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr3_+_152268920 | 0.23 |
ENST00000495875.6
ENST00000324210.10 ENST00000493459.5 |
MBNL1
|
muscleblind like splicing regulator 1 |
| chr1_-_21176836 | 0.23 |
ENST00000634879.2
ENST00000400422.6 ENST00000602326.5 ENST00000411888.5 ENST00000438975.5 ENST00000374935.7 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma 3 |
| chr17_-_42136431 | 0.22 |
ENST00000552162.5
ENST00000550504.5 |
RAB5C
|
RAB5C, member RAS oncogene family |
| chr11_+_114060204 | 0.22 |
ENST00000683318.1
|
ZBTB16
|
zinc finger and BTB domain containing 16 |
| chrX_-_16712572 | 0.22 |
ENST00000359276.9
|
CTPS2
|
CTP synthase 2 |
| chr11_-_44950867 | 0.22 |
ENST00000528290.5
ENST00000525680.6 ENST00000530035.5 ENST00000527685.5 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr6_+_32969165 | 0.22 |
ENST00000496118.2
ENST00000449085.4 |
BRD2
|
bromodomain containing 2 |
| chr5_+_141350081 | 0.22 |
ENST00000523390.2
ENST00000611598.1 |
PCDHGB1
|
protocadherin gamma subfamily B, 1 |
| chr14_-_34874887 | 0.22 |
ENST00000382422.6
|
BAZ1A
|
bromodomain adjacent to zinc finger domain 1A |
| chr2_-_70248598 | 0.22 |
ENST00000445587.5
ENST00000433529.7 ENST00000415783.6 |
TIA1
|
TIA1 cytotoxic granule associated RNA binding protein |
| chr21_-_39313578 | 0.21 |
ENST00000380800.7
|
BRWD1
|
bromodomain and WD repeat domain containing 1 |
| chr1_+_33256479 | 0.21 |
ENST00000539719.6
ENST00000483388.5 |
ZNF362
|
zinc finger protein 362 |
| chr11_-_44950839 | 0.21 |
ENST00000395648.7
ENST00000531928.6 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr22_-_38084093 | 0.21 |
ENST00000681075.1
|
SLC16A8
|
solute carrier family 16 member 8 |
| chr2_+_61065863 | 0.21 |
ENST00000402291.6
|
KIAA1841
|
KIAA1841 |
| chr11_+_46381033 | 0.21 |
ENST00000359803.7
|
MDK
|
midkine |
| chr2_-_86563382 | 0.21 |
ENST00000263856.9
|
CHMP3
|
charged multivesicular body protein 3 |
| chr19_-_17460804 | 0.21 |
ENST00000594663.1
|
ENSG00000269035.1
|
novel protein |
| chr10_+_22316375 | 0.20 |
ENST00000376836.8
ENST00000456711.5 ENST00000444869.5 ENST00000475460.6 ENST00000602390.5 ENST00000489125.2 |
COMMD3
COMMD3-BMI1
|
COMM domain containing 3 COMMD3-BMI1 readthrough |
| chr7_+_80638633 | 0.20 |
ENST00000447544.7
ENST00000482059.6 |
CD36
|
CD36 molecule |
| chr17_-_78717018 | 0.20 |
ENST00000585509.5
|
CYTH1
|
cytohesin 1 |
| chr8_+_11802667 | 0.20 |
ENST00000443614.6
ENST00000220584.9 ENST00000525900.5 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr11_+_73308237 | 0.20 |
ENST00000263674.4
|
ARHGEF17
|
Rho guanine nucleotide exchange factor 17 |
| chr19_+_48469354 | 0.20 |
ENST00000452733.7
ENST00000641098.1 |
CYTH2
|
cytohesin 2 |
| chr1_+_150067279 | 0.20 |
ENST00000643970.1
ENST00000535106.5 ENST00000369128.9 |
VPS45
|
vacuolar protein sorting 45 homolog |
| chr12_+_59664677 | 0.19 |
ENST00000548610.5
|
SLC16A7
|
solute carrier family 16 member 7 |
| chr5_+_102808057 | 0.19 |
ENST00000684043.1
ENST00000682407.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chrX_-_40096190 | 0.19 |
ENST00000679513.1
|
BCOR
|
BCL6 corepressor |
| chr8_+_11802611 | 0.19 |
ENST00000623368.3
|
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr11_+_46381194 | 0.19 |
ENST00000533952.5
|
MDK
|
midkine |
| chr12_+_22625357 | 0.19 |
ENST00000545979.2
|
ETNK1
|
ethanolamine kinase 1 |
| chr10_+_22316445 | 0.19 |
ENST00000448361.5
|
COMMD3
|
COMM domain containing 3 |
| chr1_+_93448155 | 0.19 |
ENST00000370253.6
|
FNBP1L
|
formin binding protein 1 like |
| chr12_+_69585666 | 0.19 |
ENST00000543146.2
|
CCT2
|
chaperonin containing TCP1 subunit 2 |
| chr22_+_38656627 | 0.19 |
ENST00000411557.5
ENST00000396811.6 ENST00000216029.7 ENST00000416285.5 |
CBY1
|
chibby family member 1, beta catenin antagonist |
| chr2_-_157325808 | 0.19 |
ENST00000410096.6
ENST00000420719.6 ENST00000409216.5 ENST00000419116.2 |
ERMN
|
ermin |
| chr5_-_157575767 | 0.19 |
ENST00000257527.9
|
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr10_+_21524627 | 0.18 |
ENST00000651097.1
|
MLLT10
|
MLLT10 histone lysine methyltransferase DOT1L cofactor |
| chr2_-_70248454 | 0.18 |
ENST00000416149.6
ENST00000282574.8 |
TIA1
|
TIA1 cytotoxic granule associated RNA binding protein |
| chr1_-_201507116 | 0.18 |
ENST00000340006.7
ENST00000526256.5 ENST00000526723.5 ENST00000524951.5 |
CSRP1
|
cysteine and glycine rich protein 1 |
| chr11_-_95232514 | 0.18 |
ENST00000634898.1
ENST00000542176.1 ENST00000278499.6 |
SESN3
|
sestrin 3 |
| chr17_+_77451244 | 0.18 |
ENST00000591088.5
|
SEPTIN9
|
septin 9 |
| chr7_-_108003122 | 0.18 |
ENST00000393559.2
ENST00000222399.11 ENST00000676777.1 ENST00000439976.6 ENST00000393560.5 ENST00000677793.1 ENST00000679244.1 |
LAMB1
|
laminin subunit beta 1 |
| chr2_+_69013379 | 0.18 |
ENST00000409349.7
|
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr16_+_2033264 | 0.18 |
ENST00000565855.5
ENST00000566198.1 |
SLC9A3R2
|
SLC9A3 regulator 2 |
| chr14_-_34875348 | 0.17 |
ENST00000360310.6
|
BAZ1A
|
bromodomain adjacent to zinc finger domain 1A |
| chrX_+_130339886 | 0.17 |
ENST00000543953.5
ENST00000612248.4 ENST00000424447.5 ENST00000545805.6 |
SLC25A14
|
solute carrier family 25 member 14 |
| chr15_+_51751587 | 0.17 |
ENST00000539962.6
ENST00000249700.9 |
TMOD2
|
tropomodulin 2 |
| chr11_+_60924452 | 0.17 |
ENST00000453848.7
ENST00000544065.5 ENST00000005286.8 |
TMEM132A
|
transmembrane protein 132A |
| chr2_+_10122730 | 0.17 |
ENST00000304567.10
|
RRM2
|
ribonucleotide reductase regulatory subunit M2 |
| chr21_+_37073213 | 0.16 |
ENST00000418766.5
ENST00000450533.5 ENST00000438055.5 ENST00000355666.5 ENST00000540756.5 ENST00000399010.5 |
TTC3
|
tetratricopeptide repeat domain 3 |
| chr9_+_136980211 | 0.16 |
ENST00000444903.2
|
PTGDS
|
prostaglandin D2 synthase |
| chr14_+_23322019 | 0.16 |
ENST00000557702.5
|
PABPN1
|
poly(A) binding protein nuclear 1 |
| chr3_+_136022734 | 0.15 |
ENST00000334546.6
|
PPP2R3A
|
protein phosphatase 2 regulatory subunit B''alpha |
| chr5_+_141177790 | 0.15 |
ENST00000239444.4
ENST00000623995.1 |
PCDHB8
ENSG00000279472.1
|
protocadherin beta 8 novel transcript |
| chr9_-_39239174 | 0.15 |
ENST00000358144.6
|
CNTNAP3
|
contactin associated protein family member 3 |
| chr3_-_197298092 | 0.15 |
ENST00000392382.6
|
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr12_-_85836372 | 0.15 |
ENST00000361228.5
|
RASSF9
|
Ras association domain family member 9 |
| chr12_+_50504970 | 0.15 |
ENST00000301180.10
|
DIP2B
|
disco interacting protein 2 homolog B |
| chr20_-_17531366 | 0.15 |
ENST00000377873.8
|
BFSP1
|
beaded filament structural protein 1 |
| chr6_+_87344812 | 0.14 |
ENST00000388923.5
|
C6orf163
|
chromosome 6 open reading frame 163 |
| chr3_+_133400046 | 0.14 |
ENST00000302334.3
|
BFSP2
|
beaded filament structural protein 2 |
| chr12_+_22625182 | 0.14 |
ENST00000538218.2
|
ETNK1
|
ethanolamine kinase 1 |
| chr7_+_77538059 | 0.14 |
ENST00000435495.6
|
PTPN12
|
protein tyrosine phosphatase non-receptor type 12 |
| chr7_-_19773569 | 0.14 |
ENST00000422233.5
ENST00000433641.5 ENST00000405844.6 |
TMEM196
|
transmembrane protein 196 |
| chr8_-_119673368 | 0.14 |
ENST00000427067.6
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr15_-_72783685 | 0.14 |
ENST00000456471.3
ENST00000311669.12 |
ADPGK
|
ADP dependent glucokinase |
| chr16_-_31073712 | 0.14 |
ENST00000414399.1
|
ZNF668
|
zinc finger protein 668 |
| chrX_+_130339941 | 0.14 |
ENST00000218197.9
|
SLC25A14
|
solute carrier family 25 member 14 |
| chr10_+_21524670 | 0.13 |
ENST00000631589.1
|
MLLT10
|
MLLT10 histone lysine methyltransferase DOT1L cofactor |
| chr4_+_53377630 | 0.13 |
ENST00000337488.11
ENST00000358575.9 ENST00000507922.5 ENST00000306932.10 |
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chrX_-_2500555 | 0.13 |
ENST00000381218.8
|
ZBED1
|
zinc finger BED-type containing 1 |
| chr11_-_62646598 | 0.13 |
ENST00000648273.1
ENST00000356638.8 ENST00000346178.8 ENST00000534779.5 ENST00000525994.1 ENST00000540933.5 |
GANAB
|
glucosidase II alpha subunit |
| chr8_-_33567118 | 0.13 |
ENST00000256257.2
|
RNF122
|
ring finger protein 122 |
| chr16_-_1782526 | 0.12 |
ENST00000566339.6
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chrX_+_146809768 | 0.12 |
ENST00000438525.3
|
CXorf51B
|
chromosome X open reading frame 51B |
| chr9_+_131228109 | 0.12 |
ENST00000498010.2
ENST00000476004.5 ENST00000528406.1 |
NUP214
|
nucleoporin 214 |
| chr6_+_31815532 | 0.12 |
ENST00000375651.7
ENST00000608703.1 |
HSPA1A
|
heat shock protein family A (Hsp70) member 1A |
| chr20_-_35954461 | 0.12 |
ENST00000305978.7
|
SCAND1
|
SCAN domain containing 1 |
| chr2_+_25042064 | 0.12 |
ENST00000403714.8
ENST00000401432.7 |
EFR3B
|
EFR3 homolog B |
| chr3_-_15521675 | 0.12 |
ENST00000383788.10
ENST00000383786.9 ENST00000603808.5 |
COLQ
|
collagen like tail subunit of asymmetric acetylcholinesterase |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0006683 | galactosylceramide catabolic process(GO:0006683) |
| 0.4 | 1.3 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.3 | 1.0 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.3 | 1.0 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.3 | 1.1 | GO:0021571 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.3 | 0.8 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.2 | 1.1 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.2 | 0.6 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.2 | 2.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.2 | 0.7 | GO:0050904 | diapedesis(GO:0050904) glomerular endothelium development(GO:0072011) |
| 0.2 | 0.5 | GO:1904719 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.2 | 0.6 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.1 | 0.4 | GO:0051040 | regulation of calcium-independent cell-cell adhesion(GO:0051040) corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.1 | 2.0 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.4 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.1 | 1.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 1.4 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 0.3 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.1 | 0.4 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.1 | 0.2 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.1 | 0.5 | GO:0035549 | positive regulation of interferon-beta secretion(GO:0035549) |
| 0.1 | 2.9 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 0.4 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.1 | 0.4 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.5 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.7 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 | 0.4 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 0.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.5 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.2 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 0.2 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.2 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.2 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.4 | GO:2000334 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.3 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.4 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.1 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.0 | 0.1 | GO:0001507 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.0 | 0.3 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.3 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.4 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.2 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.0 | 0.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.3 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.0 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.2 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.4 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.3 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.1 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.0 | 0.1 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.0 | GO:0003290 | atrial septum secundum morphogenesis(GO:0003290) |
| 0.0 | 0.6 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.1 | GO:0034239 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.0 | 0.2 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 5.8 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.3 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.1 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.0 | 0.5 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.2 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.1 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.4 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 0.9 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 0.4 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.1 | 0.5 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 0.3 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 0.2 | GO:0005606 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 0.0 | 2.0 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0004336 | galactosylceramidase activity(GO:0004336) |
| 0.2 | 0.5 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.1 | 0.5 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 2.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.7 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.1 | 0.4 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.1 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.4 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.1 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.4 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.3 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 0.7 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.8 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 1.0 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.4 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.0 | 0.7 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.6 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.3 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.3 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 1.4 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.0 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.0 | 2.2 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.0 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.0 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.0 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.0 | 1.1 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.4 | GO:0008143 | poly(A) binding(GO:0008143) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 2.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 1.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.6 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.6 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.7 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 1.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 2.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.1 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 1.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |