Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for SOX7
Z-value: 0.34
Transcription factors associated with SOX7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX7
|
ENSG00000171056.8 | SOX7 |
|
PINX1
|
ENSG00000258724.1 | PINX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PINX1 | hg38_v1_chr8_-_10839818_10839855 | -0.50 | 1.2e-02 | Click! |
| SOX7 | hg38_v1_chr8_-_10730498_10730525 | -0.48 | 1.5e-02 | Click! |
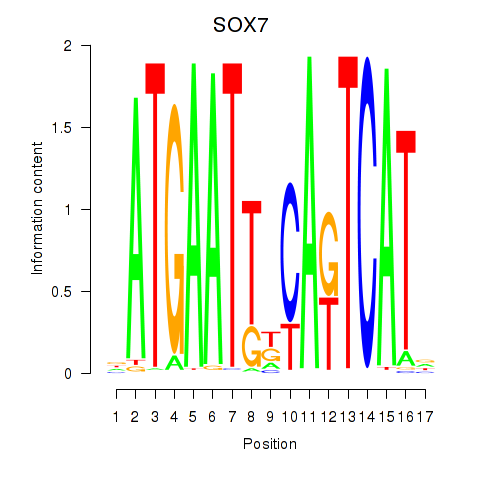
Activity profile of SOX7 motif
Sorted Z-values of SOX7 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX7
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_35880350 | 1.10 |
ENST00000605140.6
ENST00000651122.1 ENST00000603197.6 |
CCL5
|
C-C motif chemokine ligand 5 |
| chr12_-_10998304 | 0.55 |
ENST00000538986.2
|
TAS2R20
|
taste 2 receptor member 20 |
| chr4_+_76074701 | 0.34 |
ENST00000355810.9
ENST00000349321.7 |
ART3
|
ADP-ribosyltransferase 3 (inactive) |
| chr6_+_116399395 | 0.30 |
ENST00000644499.1
|
ENSG00000285446.1
|
novel protein |
| chr5_+_162067764 | 0.30 |
ENST00000639213.2
ENST00000414552.6 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr12_-_89656093 | 0.29 |
ENST00000359142.7
|
ATP2B1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr3_-_122564577 | 0.29 |
ENST00000477522.6
ENST00000360356.6 |
PARP9
|
poly(ADP-ribose) polymerase family member 9 |
| chr1_+_222928415 | 0.28 |
ENST00000284476.7
|
DISP1
|
dispatched RND transporter family member 1 |
| chr5_+_162067858 | 0.28 |
ENST00000361925.9
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr5_+_162067500 | 0.27 |
ENST00000639384.1
ENST00000640985.1 ENST00000638772.1 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr12_-_89656051 | 0.27 |
ENST00000261173.6
|
ATP2B1
|
ATPase plasma membrane Ca2+ transporting 1 |
| chr1_-_158426237 | 0.27 |
ENST00000641042.1
|
OR10K2
|
olfactory receptor family 10 subfamily K member 2 |
| chr10_+_94683771 | 0.25 |
ENST00000339022.6
|
CYP2C18
|
cytochrome P450 family 2 subfamily C member 18 |
| chr5_+_162067458 | 0.25 |
ENST00000639975.1
ENST00000639111.2 ENST00000639683.1 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr19_+_34926892 | 0.25 |
ENST00000303586.11
ENST00000601142.2 ENST00000439785.5 ENST00000601540.5 ENST00000601957.5 |
ZNF30
|
zinc finger protein 30 |
| chr6_+_143843316 | 0.23 |
ENST00000367576.6
|
LTV1
|
LTV1 ribosome biogenesis factor |
| chr7_-_138755892 | 0.22 |
ENST00000644341.1
ENST00000478480.2 |
ATP6V0A4
|
ATPase H+ transporting V0 subunit a4 |
| chr3_+_107599309 | 0.22 |
ENST00000406780.5
|
BBX
|
BBX high mobility group box domain containing |
| chr16_-_20352707 | 0.21 |
ENST00000396134.6
ENST00000573567.5 ENST00000570757.5 ENST00000396138.9 ENST00000571174.5 ENST00000576688.2 |
UMOD
|
uromodulin |
| chr4_+_74308463 | 0.20 |
ENST00000413830.6
|
EPGN
|
epithelial mitogen |
| chr13_-_37059432 | 0.20 |
ENST00000464744.5
|
SUPT20H
|
SPT20 homolog, SAGA complex component |
| chr13_+_94601830 | 0.20 |
ENST00000376958.5
|
GPR180
|
G protein-coupled receptor 180 |
| chr4_-_122456725 | 0.20 |
ENST00000226730.5
|
IL2
|
interleukin 2 |
| chr6_-_111483700 | 0.18 |
ENST00000435970.5
ENST00000358835.7 |
REV3L
|
REV3 like, DNA directed polymerase zeta catalytic subunit |
| chr12_+_63779894 | 0.18 |
ENST00000261234.11
|
RXYLT1
|
ribitol xylosyltransferase 1 |
| chr18_+_35241027 | 0.17 |
ENST00000330501.12
ENST00000601719.1 ENST00000591206.5 ENST00000261333.10 ENST00000355632.8 ENST00000585800.1 |
ZNF397
|
zinc finger protein 397 |
| chr1_+_100133135 | 0.17 |
ENST00000370143.5
ENST00000370141.7 |
TRMT13
|
tRNA methyltransferase 13 homolog |
| chr4_-_47981535 | 0.15 |
ENST00000402813.9
|
CNGA1
|
cyclic nucleotide gated channel subunit alpha 1 |
| chr11_+_7597182 | 0.13 |
ENST00000528883.5
|
PPFIBP2
|
PPFIA binding protein 2 |
| chr4_+_68447453 | 0.11 |
ENST00000305363.9
|
TMPRSS11E
|
transmembrane serine protease 11E |
| chr10_-_12042771 | 0.10 |
ENST00000357604.10
|
UPF2
|
UPF2 regulator of nonsense mediated mRNA decay |
| chr1_-_206003385 | 0.10 |
ENST00000617070.5
|
RAB7B
|
RAB7B, member RAS oncogene family |
| chr11_+_35186820 | 0.09 |
ENST00000531110.6
ENST00000525685.6 |
CD44
|
CD44 molecule (Indian blood group) |
| chr16_+_28292485 | 0.09 |
ENST00000341901.5
|
SBK1
|
SH3 domain binding kinase 1 |
| chr3_-_36993103 | 0.09 |
ENST00000322716.8
|
EPM2AIP1
|
EPM2A interacting protein 1 |
| chr14_-_106117159 | 0.08 |
ENST00000390601.3
|
IGHV3-11
|
immunoglobulin heavy variable 3-11 |
| chr21_-_14546297 | 0.08 |
ENST00000400566.6
ENST00000400564.5 |
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr6_+_42563981 | 0.08 |
ENST00000372899.6
ENST00000372901.2 |
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr1_-_206003442 | 0.08 |
ENST00000623893.1
|
RAB7B
|
RAB7B, member RAS oncogene family |
| chrX_+_131083706 | 0.06 |
ENST00000370921.1
|
ARHGAP36
|
Rho GTPase activating protein 36 |
| chr8_-_86069662 | 0.06 |
ENST00000276616.3
|
PSKH2
|
protein serine kinase H2 |
| chr8_+_28338640 | 0.06 |
ENST00000522209.1
|
PNOC
|
prepronociceptin |
| chr10_+_4963406 | 0.04 |
ENST00000380872.9
ENST00000442997.5 |
AKR1C1
|
aldo-keto reductase family 1 member C1 |
| chr5_+_141182369 | 0.03 |
ENST00000609684.3
ENST00000625044.1 ENST00000623407.1 ENST00000623884.1 |
PCDHB16
ENSG00000279068.1
|
protocadherin beta 16 novel transcript |
| chr16_+_33009175 | 0.02 |
ENST00000565407.2
|
IGHV3OR16-8
|
immunoglobulin heavy variable 3/OR16-8 (non-functional) |
| chr15_+_43826961 | 0.02 |
ENST00000381246.6
ENST00000452115.1 ENST00000263795.11 |
WDR76
|
WD repeat domain 76 |
| chr19_-_38426195 | 0.02 |
ENST00000615439.5
ENST00000614135.4 ENST00000622174.4 ENST00000587753.5 ENST00000454404.6 ENST00000617966.4 ENST00000618320.4 ENST00000293062.13 ENST00000433821.6 ENST00000426920.6 |
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr17_-_41467386 | 0.02 |
ENST00000225899.4
|
KRT32
|
keratin 32 |
| chrX_-_135781729 | 0.01 |
ENST00000617203.1
|
CT45A5
|
cancer/testis antigen family 45 member A5 |
| chr11_-_62707413 | 0.01 |
ENST00000360796.10
ENST00000449636.6 |
BSCL2
|
BSCL2 lipid droplet biogenesis associated, seipin |
| chr3_+_98463201 | 0.01 |
ENST00000642057.1
|
OR5K1
|
olfactory receptor family 5 subfamily K member 1 |
| chr11_+_56176618 | 0.01 |
ENST00000312298.1
|
OR5J2
|
olfactory receptor family 5 subfamily J member 2 |
| chr16_-_33845229 | 0.00 |
ENST00000569103.2
|
IGHV3OR16-17
|
immunoglobulin heavy variable 3/OR16-17 (non-functional) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.3 | GO:0035638 | patched ligand maturation(GO:0007225) signal maturation(GO:0035638) |
| 0.1 | 0.2 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.1 | 0.6 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.0 | 0.2 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 1.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.3 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.2 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.2 | GO:0034448 | EGO complex(GO:0034448) |
| 0.0 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0031726 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 1.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.3 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.6 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.0 | 0.0 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.3 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.3 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |