Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
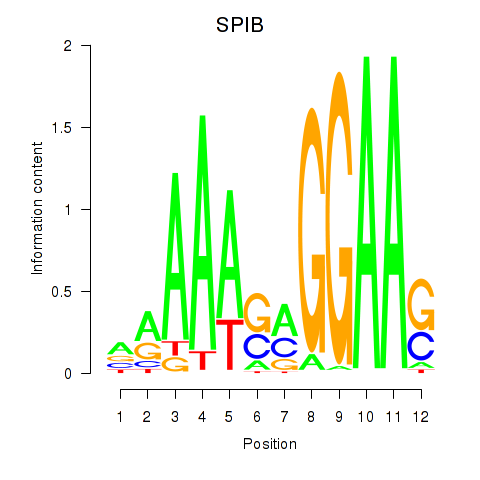
Results for SPIB
Z-value: 0.84
Transcription factors associated with SPIB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SPIB
|
ENSG00000269404.7 | SPIB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SPIB | hg38_v1_chr19_+_50418930_50418958 | 0.04 | 8.6e-01 | Click! |
Activity profile of SPIB motif
Sorted Z-values of SPIB motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SPIB
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_135851681 | 1.51 |
ENST00000308191.11
|
PDE7B
|
phosphodiesterase 7B |
| chr2_-_224401994 | 1.46 |
ENST00000389874.3
|
FAM124B
|
family with sequence similarity 124 member B |
| chr2_-_224402097 | 1.45 |
ENST00000409685.4
|
FAM124B
|
family with sequence similarity 124 member B |
| chr19_-_13102848 | 1.37 |
ENST00000264824.5
|
LYL1
|
LYL1 basic helix-loop-helix family member |
| chr13_+_57631735 | 1.30 |
ENST00000377918.8
|
PCDH17
|
protocadherin 17 |
| chr22_+_19760714 | 1.25 |
ENST00000649276.2
|
TBX1
|
T-box transcription factor 1 |
| chr19_+_45469841 | 1.08 |
ENST00000592811.5
ENST00000586615.5 |
FOSB
|
FosB proto-oncogene, AP-1 transcription factor subunit |
| chr1_-_121183911 | 1.06 |
ENST00000355228.8
|
FAM72B
|
family with sequence similarity 72 member B |
| chr14_+_73950489 | 1.06 |
ENST00000554320.1
|
COQ6
|
coenzyme Q6, monooxygenase |
| chr19_+_2476118 | 1.05 |
ENST00000215631.9
ENST00000587345.1 |
GADD45B
|
growth arrest and DNA damage inducible beta |
| chr16_+_30949054 | 1.03 |
ENST00000318663.5
ENST00000566237.1 ENST00000562699.1 |
ORAI3
|
ORAI calcium release-activated calcium modulator 3 |
| chr1_-_206202419 | 1.00 |
ENST00000607379.1
ENST00000341209.9 |
FAM72A
|
family with sequence similarity 72 member A |
| chr16_-_4800470 | 1.00 |
ENST00000592019.1
ENST00000586153.1 |
ROGDI
|
rogdi atypical leucine zipper |
| chr2_+_233059838 | 0.97 |
ENST00000359570.9
|
INPP5D
|
inositol polyphosphate-5-phosphatase D |
| chr2_+_233060295 | 0.97 |
ENST00000445964.6
|
INPP5D
|
inositol polyphosphate-5-phosphatase D |
| chr17_-_74712911 | 0.94 |
ENST00000326165.11
ENST00000583937.5 ENST00000301573.13 ENST00000469092.5 |
CD300LF
|
CD300 molecule like family member f |
| chr8_-_77000194 | 0.89 |
ENST00000522527.5
|
PEX2
|
peroxisomal biogenesis factor 2 |
| chr7_+_74773962 | 0.74 |
ENST00000289473.10
|
NCF1
|
neutrophil cytosolic factor 1 |
| chr1_-_153946652 | 0.73 |
ENST00000361217.9
|
DENND4B
|
DENN domain containing 4B |
| chr19_-_17264718 | 0.73 |
ENST00000431146.6
ENST00000594190.5 |
USHBP1
|
USH1 protein network component harmonin binding protein 1 |
| chr1_+_159302321 | 0.69 |
ENST00000368114.1
|
FCER1A
|
Fc fragment of IgE receptor Ia |
| chr22_-_37519528 | 0.68 |
ENST00000403299.5
|
CARD10
|
caspase recruitment domain family member 10 |
| chr22_-_37519349 | 0.68 |
ENST00000251973.10
|
CARD10
|
caspase recruitment domain family member 10 |
| chr17_-_19046957 | 0.65 |
ENST00000284154.10
ENST00000573099.5 |
GRAP
|
GRB2 related adaptor protein |
| chr13_+_30735523 | 0.64 |
ENST00000380490.5
|
ALOX5AP
|
arachidonate 5-lipoxygenase activating protein |
| chr13_+_30713477 | 0.63 |
ENST00000617770.4
|
ALOX5AP
|
arachidonate 5-lipoxygenase activating protein |
| chrX_-_75156272 | 0.63 |
ENST00000620875.5
ENST00000669573.1 ENST00000339447.8 ENST00000645829.3 ENST00000529949.5 ENST00000373394.8 ENST00000253577.9 ENST00000644766.1 ENST00000534524.5 |
ABCB7
|
ATP binding cassette subfamily B member 7 |
| chr12_-_14961256 | 0.62 |
ENST00000541380.5
|
ARHGDIB
|
Rho GDP dissociation inhibitor beta |
| chr17_-_31314066 | 0.62 |
ENST00000577894.1
|
EVI2B
|
ecotropic viral integration site 2B |
| chr1_-_121184292 | 0.62 |
ENST00000452190.2
ENST00000619376.4 ENST00000369390.7 |
FAM72B
|
family with sequence similarity 72 member B |
| chr6_-_109094819 | 0.61 |
ENST00000436639.6
|
SESN1
|
sestrin 1 |
| chr3_-_116444983 | 0.61 |
ENST00000333617.8
|
LSAMP
|
limbic system associated membrane protein |
| chr1_-_143971965 | 0.61 |
ENST00000369175.4
ENST00000584486.6 |
FAM72C
|
family with sequence similarity 72 member C |
| chr7_-_36724543 | 0.60 |
ENST00000612871.4
|
AOAH
|
acyloxyacyl hydrolase |
| chr7_-_36724380 | 0.58 |
ENST00000617267.4
|
AOAH
|
acyloxyacyl hydrolase |
| chr17_+_7558296 | 0.58 |
ENST00000438470.5
ENST00000436057.5 |
TNFSF13
|
TNF superfamily member 13 |
| chr11_-_64803152 | 0.58 |
ENST00000439069.5
ENST00000294066.7 ENST00000377350.7 |
MAP4K2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr8_+_73991345 | 0.57 |
ENST00000284818.7
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr11_+_64340191 | 0.56 |
ENST00000356786.10
|
CCDC88B
|
coiled-coil domain containing 88B |
| chr7_-_36724457 | 0.56 |
ENST00000617537.5
ENST00000435386.1 |
AOAH
|
acyloxyacyl hydrolase |
| chr13_+_108269880 | 0.55 |
ENST00000542136.1
|
TNFSF13B
|
TNF superfamily member 13b |
| chr7_-_138755892 | 0.52 |
ENST00000644341.1
ENST00000478480.2 |
ATP6V0A4
|
ATPase H+ transporting V0 subunit a4 |
| chr6_-_132513045 | 0.52 |
ENST00000367941.7
ENST00000367937.4 |
STX7
|
syntaxin 7 |
| chr1_-_206202827 | 0.51 |
ENST00000431655.2
ENST00000367128.8 |
FAM72A
|
family with sequence similarity 72 member A |
| chr14_+_66824439 | 0.51 |
ENST00000555456.1
|
GPHN
|
gephyrin |
| chr11_+_47269363 | 0.49 |
ENST00000342922.8
|
MADD
|
MAP kinase activating death domain |
| chr8_+_124973288 | 0.49 |
ENST00000319286.6
|
ZNF572
|
zinc finger protein 572 |
| chr1_+_145095967 | 0.49 |
ENST00000400889.3
|
FAM72D
|
family with sequence similarity 72 member D |
| chr9_-_21995262 | 0.47 |
ENST00000494262.5
|
CDKN2A
|
cyclin dependent kinase inhibitor 2A |
| chr12_-_27014300 | 0.46 |
ENST00000535819.1
ENST00000543803.5 ENST00000535423.5 ENST00000539741.5 ENST00000343028.9 ENST00000545600.1 ENST00000543088.5 |
TM7SF3
|
transmembrane 7 superfamily member 3 |
| chr9_+_101533840 | 0.46 |
ENST00000389120.8
ENST00000374819.6 ENST00000479306.5 |
RNF20
|
ring finger protein 20 |
| chr16_+_54930827 | 0.45 |
ENST00000394636.9
|
IRX5
|
iroquois homeobox 5 |
| chr9_+_127611760 | 0.45 |
ENST00000625363.2
ENST00000626539.3 |
STXBP1
|
syntaxin binding protein 1 |
| chr17_-_5234801 | 0.45 |
ENST00000571800.5
ENST00000574081.6 ENST00000399600.8 ENST00000574297.1 |
SCIMP
|
SLP adaptor and CSK interacting membrane protein |
| chr9_+_127612222 | 0.44 |
ENST00000637953.1
ENST00000636962.2 |
STXBP1
|
syntaxin binding protein 1 |
| chr16_+_57542635 | 0.44 |
ENST00000349457.8
|
ADGRG5
|
adhesion G protein-coupled receptor G5 |
| chr16_+_57542672 | 0.44 |
ENST00000615867.4
ENST00000340339.4 |
ADGRG5
|
adhesion G protein-coupled receptor G5 |
| chr6_-_144064511 | 0.44 |
ENST00000626373.2
ENST00000628651.2 ENST00000626294.2 ENST00000437412.5 ENST00000444202.5 ENST00000626462.2 ENST00000627449.2 |
PLAGL1
|
PLAG1 like zinc finger 1 |
| chr9_+_127612257 | 0.43 |
ENST00000637173.2
ENST00000630492.2 ENST00000627871.2 ENST00000373302.8 ENST00000373299.5 ENST00000650920.1 ENST00000476182.3 |
STXBP1
|
syntaxin binding protein 1 |
| chr19_-_38426162 | 0.43 |
ENST00000587738.2
ENST00000586305.5 |
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr19_-_48363914 | 0.43 |
ENST00000377431.6
ENST00000293261.8 |
TMEM143
|
transmembrane protein 143 |
| chr2_+_96326204 | 0.43 |
ENST00000420728.1
ENST00000361124.5 |
ITPRIPL1
|
ITPRIP like 1 |
| chr9_-_96383675 | 0.42 |
ENST00000375257.2
ENST00000375259.9 ENST00000253270.13 |
SLC35D2
|
solute carrier family 35 member D2 |
| chr6_+_15246054 | 0.42 |
ENST00000341776.7
|
JARID2
|
jumonji and AT-rich interaction domain containing 2 |
| chr19_-_48364034 | 0.42 |
ENST00000435956.7
|
TMEM143
|
transmembrane protein 143 |
| chr4_-_84966637 | 0.42 |
ENST00000295888.9
|
WDFY3
|
WD repeat and FYVE domain containing 3 |
| chr19_+_41877267 | 0.41 |
ENST00000221972.8
ENST00000597454.1 ENST00000444740.2 |
CD79A
|
CD79a molecule |
| chr16_+_30183595 | 0.40 |
ENST00000219150.10
ENST00000570045.5 ENST00000565497.5 ENST00000570244.5 |
CORO1A
|
coronin 1A |
| chr6_+_130018565 | 0.40 |
ENST00000361794.7
ENST00000526087.5 ENST00000533560.5 |
L3MBTL3
|
L3MBTL histone methyl-lysine binding protein 3 |
| chr9_+_117704168 | 0.40 |
ENST00000472304.2
ENST00000394487.5 |
TLR4
|
toll like receptor 4 |
| chr15_-_33068143 | 0.40 |
ENST00000558197.1
|
FMN1
|
formin 1 |
| chr19_-_17264732 | 0.39 |
ENST00000252597.8
|
USHBP1
|
USH1 protein network component harmonin binding protein 1 |
| chr11_+_123358416 | 0.39 |
ENST00000638157.1
|
GRAMD1B
|
GRAM domain containing 1B |
| chr9_+_132978651 | 0.38 |
ENST00000636137.1
|
GFI1B
|
growth factor independent 1B transcriptional repressor |
| chr9_-_21995301 | 0.38 |
ENST00000498628.6
|
CDKN2A
|
cyclin dependent kinase inhibitor 2A |
| chr9_+_2017383 | 0.37 |
ENST00000382194.6
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr14_+_65412949 | 0.37 |
ENST00000674118.1
ENST00000553924.5 ENST00000358307.6 ENST00000557338.5 ENST00000554610.1 |
FUT8
|
fucosyltransferase 8 |
| chr5_-_173328407 | 0.37 |
ENST00000265087.9
|
STC2
|
stanniocalcin 2 |
| chr2_-_191014137 | 0.37 |
ENST00000673777.1
ENST00000673942.1 ENST00000424722.6 ENST00000392322.7 ENST00000361099.8 ENST00000392323.6 ENST00000673816.1 ENST00000673847.1 ENST00000673952.1 ENST00000540176.6 ENST00000673841.1 |
STAT1
|
signal transducer and activator of transcription 1 |
| chr2_+_68365274 | 0.36 |
ENST00000234313.8
|
PLEK
|
pleckstrin |
| chrX_+_110003095 | 0.36 |
ENST00000372073.5
ENST00000372068.7 ENST00000288381.4 |
TMEM164
|
transmembrane protein 164 |
| chr12_-_14961610 | 0.36 |
ENST00000542276.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor beta |
| chr12_+_25052512 | 0.36 |
ENST00000557489.5
ENST00000354454.7 ENST00000536173.5 |
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr1_+_161581339 | 0.36 |
ENST00000543859.5
ENST00000611236.1 |
FCGR2C
|
Fc fragment of IgG receptor IIc (gene/pseudogene) |
| chr17_-_4786433 | 0.36 |
ENST00000354194.4
|
VMO1
|
vitelline membrane outer layer 1 homolog |
| chr9_+_2017572 | 0.36 |
ENST00000637806.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_+_108621260 | 0.35 |
ENST00000409441.5
|
LIMS1
|
LIM zinc finger domain containing 1 |
| chr6_-_154356735 | 0.35 |
ENST00000367220.8
ENST00000265198.8 ENST00000520261.1 |
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr7_-_13989658 | 0.35 |
ENST00000430479.6
ENST00000433547.1 ENST00000405192.6 |
ETV1
|
ETS variant transcription factor 1 |
| chr19_-_31349408 | 0.34 |
ENST00000240587.5
|
TSHZ3
|
teashirt zinc finger homeobox 3 |
| chr5_-_56116946 | 0.34 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr2_+_201260496 | 0.34 |
ENST00000323492.11
|
CASP8
|
caspase 8 |
| chr19_-_38426195 | 0.34 |
ENST00000615439.5
ENST00000614135.4 ENST00000622174.4 ENST00000587753.5 ENST00000454404.6 ENST00000617966.4 ENST00000618320.4 ENST00000293062.13 ENST00000433821.6 ENST00000426920.6 |
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr6_+_49463360 | 0.34 |
ENST00000335783.4
|
CENPQ
|
centromere protein Q |
| chr20_-_20052389 | 0.34 |
ENST00000536226.2
|
CRNKL1
|
crooked neck pre-mRNA splicing factor 1 |
| chr12_+_51391273 | 0.34 |
ENST00000535225.6
ENST00000358657.7 |
SLC4A8
|
solute carrier family 4 member 8 |
| chr7_-_122886706 | 0.34 |
ENST00000313070.11
ENST00000334010.11 ENST00000615869.4 |
CADPS2
|
calcium dependent secretion activator 2 |
| chr6_+_37005630 | 0.34 |
ENST00000274963.13
|
FGD2
|
FYVE, RhoGEF and PH domain containing 2 |
| chr21_-_44920918 | 0.34 |
ENST00000522688.5
|
ITGB2
|
integrin subunit beta 2 |
| chr14_+_65412717 | 0.34 |
ENST00000673929.1
|
FUT8
|
fucosyltransferase 8 |
| chr9_+_68705230 | 0.33 |
ENST00000265382.8
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase type 1 beta |
| chr2_-_70553440 | 0.33 |
ENST00000450929.5
|
TGFA
|
transforming growth factor alpha |
| chr20_-_47786553 | 0.33 |
ENST00000467815.5
ENST00000359930.8 ENST00000484875.5 |
SULF2
|
sulfatase 2 |
| chr9_+_68705414 | 0.33 |
ENST00000541509.5
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase type 1 beta |
| chr22_-_37484505 | 0.33 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr20_-_3712564 | 0.32 |
ENST00000344754.6
|
SIGLEC1
|
sialic acid binding Ig like lectin 1 |
| chr12_-_7503744 | 0.32 |
ENST00000396620.7
ENST00000432237.3 |
CD163
|
CD163 molecule |
| chr12_-_53507482 | 0.32 |
ENST00000267017.4
ENST00000448979.4 |
NPFF
|
neuropeptide FF-amide peptide precursor |
| chr2_+_219597838 | 0.32 |
ENST00000456909.6
|
STK11IP
|
serine/threonine kinase 11 interacting protein |
| chr19_-_3786254 | 0.32 |
ENST00000585778.5
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr17_+_19127535 | 0.31 |
ENST00000577213.1
ENST00000344415.9 |
GRAPL
|
GRB2 related adaptor protein like |
| chr9_+_117704382 | 0.31 |
ENST00000646089.1
ENST00000355622.8 |
ENSG00000285082.2
TLR4
|
novel protein toll like receptor 4 |
| chr19_-_3786363 | 0.31 |
ENST00000310132.11
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr19_-_3786408 | 0.31 |
ENST00000395040.6
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr1_+_209768482 | 0.31 |
ENST00000367023.5
|
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr14_+_101761786 | 0.31 |
ENST00000422945.6
ENST00000554442.5 ENST00000556260.6 ENST00000328724.9 ENST00000557268.5 |
PPP2R5C
|
protein phosphatase 2 regulatory subunit B'gamma |
| chrX_+_70133433 | 0.30 |
ENST00000356413.5
ENST00000342206.10 |
IGBP1
|
immunoglobulin binding protein 1 |
| chr1_+_151612001 | 0.30 |
ENST00000642376.1
ENST00000368843.8 ENST00000458013.7 |
SNX27
|
sorting nexin 27 |
| chr9_-_127778659 | 0.30 |
ENST00000314830.13
|
SH2D3C
|
SH2 domain containing 3C |
| chr5_+_65563239 | 0.30 |
ENST00000535264.5
ENST00000538977.5 ENST00000261308.10 |
PPWD1
|
peptidylprolyl isomerase domain and WD repeat containing 1 |
| chr12_-_7503841 | 0.30 |
ENST00000359156.8
|
CD163
|
CD163 molecule |
| chr2_-_191013955 | 0.29 |
ENST00000409465.5
|
STAT1
|
signal transducer and activator of transcription 1 |
| chr3_-_121660892 | 0.29 |
ENST00000428394.6
ENST00000314583.8 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr2_-_151261839 | 0.28 |
ENST00000331426.6
|
RBM43
|
RNA binding motif protein 43 |
| chr12_-_14961559 | 0.28 |
ENST00000228945.9
ENST00000541546.5 |
ARHGDIB
|
Rho GDP dissociation inhibitor beta |
| chr2_+_137964279 | 0.28 |
ENST00000329366.8
|
HNMT
|
histamine N-methyltransferase |
| chr12_+_49961864 | 0.28 |
ENST00000293599.7
|
AQP5
|
aquaporin 5 |
| chr5_-_65563117 | 0.28 |
ENST00000508421.5
ENST00000510693.5 ENST00000514814.5 ENST00000396679.6 ENST00000515497.5 |
CENPK
|
centromere protein K |
| chr6_-_24935942 | 0.28 |
ENST00000645100.1
ENST00000643898.2 ENST00000613507.4 |
RIPOR2
|
RHO family interacting cell polarization regulator 2 |
| chr1_+_22653189 | 0.27 |
ENST00000432749.6
|
C1QB
|
complement C1q B chain |
| chr10_+_92831153 | 0.27 |
ENST00000672817.1
|
EXOC6
|
exocyst complex component 6 |
| chr1_+_161505412 | 0.27 |
ENST00000367972.8
|
FCGR2A
|
Fc fragment of IgG receptor IIa |
| chr21_+_33266350 | 0.27 |
ENST00000290200.7
|
IL10RB
|
interleukin 10 receptor subunit beta |
| chr6_-_32178080 | 0.27 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr10_+_17809337 | 0.27 |
ENST00000569591.3
|
MRC1
|
mannose receptor C-type 1 |
| chr6_+_130827398 | 0.26 |
ENST00000541421.2
|
SMLR1
|
small leucine rich protein 1 |
| chr5_+_169637241 | 0.26 |
ENST00000520908.7
|
DOCK2
|
dedicator of cytokinesis 2 |
| chr9_-_35563867 | 0.26 |
ENST00000399742.7
ENST00000619051.4 |
FAM166B
|
family with sequence similarity 166 member B |
| chr2_-_113235443 | 0.26 |
ENST00000465084.1
|
PAX8
|
paired box 8 |
| chr10_-_73096974 | 0.26 |
ENST00000440381.5
ENST00000263556.3 |
P4HA1
|
prolyl 4-hydroxylase subunit alpha 1 |
| chr12_+_111405861 | 0.26 |
ENST00000341259.7
|
SH2B3
|
SH2B adaptor protein 3 |
| chr1_-_30757767 | 0.26 |
ENST00000294507.4
|
LAPTM5
|
lysosomal protein transmembrane 5 |
| chr6_-_49463173 | 0.25 |
ENST00000274813.4
|
MMUT
|
methylmalonyl-CoA mutase |
| chr12_-_121802886 | 0.25 |
ENST00000545885.5
ENST00000542933.5 ENST00000428029.6 ENST00000541694.5 ENST00000536662.5 ENST00000535643.5 ENST00000541657.5 |
LINC01089
RHOF
|
long intergenic non-protein coding RNA 1089 ras homolog family member F, filopodia associated |
| chr12_+_25052634 | 0.25 |
ENST00000548766.5
|
IRAG2
|
inositol 1,4,5-triphosphate receptor associated 2 |
| chr11_+_61333204 | 0.25 |
ENST00000532173.6
ENST00000394900.8 |
TKFC
|
triokinase and FMN cyclase |
| chr7_+_106865263 | 0.24 |
ENST00000440650.6
ENST00000496166.6 ENST00000473541.5 |
PIK3CG
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma |
| chr8_+_39913881 | 0.24 |
ENST00000518237.6
|
IDO1
|
indoleamine 2,3-dioxygenase 1 |
| chr10_-_73096850 | 0.24 |
ENST00000307116.6
ENST00000373008.6 ENST00000394890.7 |
P4HA1
|
prolyl 4-hydroxylase subunit alpha 1 |
| chr17_-_40937641 | 0.24 |
ENST00000209718.8
|
KRT23
|
keratin 23 |
| chr2_+_134254065 | 0.24 |
ENST00000281923.4
|
MGAT5
|
alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase |
| chr16_-_73048104 | 0.23 |
ENST00000268489.10
|
ZFHX3
|
zinc finger homeobox 3 |
| chr16_-_30370396 | 0.23 |
ENST00000409939.8
|
TBC1D10B
|
TBC1 domain family member 10B |
| chr2_-_42792558 | 0.23 |
ENST00000431905.1
ENST00000294973.11 |
HAAO
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr18_+_13218769 | 0.23 |
ENST00000677055.1
ENST00000399848.7 |
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr7_+_139829153 | 0.23 |
ENST00000652056.1
|
TBXAS1
|
thromboxane A synthase 1 |
| chr19_-_41688167 | 0.23 |
ENST00000602225.1
|
CEACAM7
|
CEA cell adhesion molecule 7 |
| chr11_+_1839602 | 0.23 |
ENST00000617947.4
ENST00000252898.11 |
TNNI2
|
troponin I2, fast skeletal type |
| chr6_-_56843638 | 0.22 |
ENST00000421834.6
ENST00000370788.6 |
DST
|
dystonin |
| chr9_+_121268060 | 0.22 |
ENST00000373808.8
ENST00000432226.7 ENST00000449733.7 |
GSN
|
gelsolin |
| chr10_+_70478761 | 0.22 |
ENST00000263563.7
|
PALD1
|
phosphatase domain containing paladin 1 |
| chr7_+_141776674 | 0.22 |
ENST00000247881.4
|
TAS2R4
|
taste 2 receptor member 4 |
| chr7_-_13989891 | 0.21 |
ENST00000405218.6
|
ETV1
|
ETS variant transcription factor 1 |
| chr18_-_50287570 | 0.21 |
ENST00000586837.1
ENST00000412036.6 ENST00000589940.5 ENST00000587396.1 ENST00000591474.5 ENST00000285106.11 |
CXXC1
|
CXXC finger protein 1 |
| chrX_+_24054931 | 0.21 |
ENST00000253039.9
ENST00000423068.1 |
EIF2S3
|
eukaryotic translation initiation factor 2 subunit gamma |
| chr14_+_44962177 | 0.20 |
ENST00000361462.7
ENST00000361577.7 |
TOGARAM1
|
TOG array regulator of axonemal microtubules 1 |
| chr4_-_86360071 | 0.20 |
ENST00000641677.1
ENST00000639234.1 ENST00000641553.1 ENST00000641826.1 ENST00000641537.1 ENST00000395169.9 ENST00000641408.1 ENST00000638225.1 ENST00000641052.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chrX_-_47629845 | 0.20 |
ENST00000469388.1
ENST00000396992.8 ENST00000377005.6 |
CFP
|
complement factor properdin |
| chr13_+_53028806 | 0.20 |
ENST00000219022.3
|
OLFM4
|
olfactomedin 4 |
| chr17_-_40937445 | 0.20 |
ENST00000436344.7
ENST00000485751.1 |
KRT23
|
keratin 23 |
| chr12_+_80716906 | 0.19 |
ENST00000228644.4
|
MYF5
|
myogenic factor 5 |
| chr14_+_85530127 | 0.19 |
ENST00000330753.6
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr5_+_160421847 | 0.19 |
ENST00000352433.10
ENST00000517480.1 ENST00000520452.5 ENST00000393964.1 |
PTTG1
|
PTTG1 regulator of sister chromatid separation, securin |
| chr5_-_10249876 | 0.19 |
ENST00000511437.6
ENST00000280330.12 ENST00000510047.5 |
ATPSCKMT
|
ATP synthase c subunit lysine N-methyltransferase |
| chr18_-_50287816 | 0.19 |
ENST00000589548.6
ENST00000673786.1 |
CXXC1
|
CXXC finger protein 1 |
| chr2_+_137964446 | 0.19 |
ENST00000280096.5
ENST00000280097.5 |
HNMT
|
histamine N-methyltransferase |
| chr16_-_4801301 | 0.18 |
ENST00000586504.5
ENST00000649556.1 |
ROGDI
ENSG00000285952.1
|
rogdi atypical leucine zipper novel transcript |
| chr19_+_35154914 | 0.18 |
ENST00000423817.7
|
FXYD5
|
FXYD domain containing ion transport regulator 5 |
| chr19_+_16143678 | 0.18 |
ENST00000613986.4
ENST00000593031.1 |
HSH2D
|
hematopoietic SH2 domain containing |
| chr6_-_145735964 | 0.18 |
ENST00000640980.1
ENST00000639423.1 ENST00000611340.5 |
EPM2A
|
EPM2A glucan phosphatase, laforin |
| chr3_+_32391871 | 0.18 |
ENST00000465248.1
|
CMTM7
|
CKLF like MARVEL transmembrane domain containing 7 |
| chr9_-_136687380 | 0.18 |
ENST00000538402.1
ENST00000371694.7 |
AGPAT2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 |
| chr11_-_47378494 | 0.18 |
ENST00000533030.1
|
SPI1
|
Spi-1 proto-oncogene |
| chr22_+_41092869 | 0.18 |
ENST00000674155.1
|
EP300
|
E1A binding protein p300 |
| chr9_+_214843 | 0.18 |
ENST00000432829.7
|
DOCK8
|
dedicator of cytokinesis 8 |
| chr2_+_147844488 | 0.18 |
ENST00000535787.5
|
ACVR2A
|
activin A receptor type 2A |
| chr12_-_54984667 | 0.17 |
ENST00000524668.5
ENST00000533607.1 ENST00000449076.6 |
TESPA1
|
thymocyte expressed, positive selection associated 1 |
| chr3_-_143848442 | 0.17 |
ENST00000474151.1
ENST00000316549.11 |
SLC9A9
|
solute carrier family 9 member A9 |
| chr3_-_179266971 | 0.17 |
ENST00000349697.2
ENST00000497599.5 |
KCNMB3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr8_-_120445092 | 0.17 |
ENST00000518918.1
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chr10_+_11164961 | 0.17 |
ENST00000399850.7
ENST00000417956.6 |
CELF2
|
CUGBP Elav-like family member 2 |
| chr11_+_1840168 | 0.17 |
ENST00000381905.3
|
TNNI2
|
troponin I2, fast skeletal type |
| chrX_-_130165699 | 0.17 |
ENST00000676328.1
ENST00000675857.1 ENST00000675427.1 ENST00000675092.1 |
AIFM1
|
apoptosis inducing factor mitochondria associated 1 |
| chr7_-_100158679 | 0.17 |
ENST00000456769.5
ENST00000316937.8 |
TRAPPC14
|
trafficking protein particle complex 14 |
| chr13_-_110561668 | 0.17 |
ENST00000267328.5
|
RAB20
|
RAB20, member RAS oncogene family |
| chrX_-_130165825 | 0.16 |
ENST00000675240.1
ENST00000319908.8 ENST00000674546.1 ENST00000287295.8 |
AIFM1
|
apoptosis inducing factor mitochondria associated 1 |
| chr9_-_136687422 | 0.16 |
ENST00000371696.7
|
AGPAT2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 |
| chr14_+_101809855 | 0.16 |
ENST00000557714.1
ENST00000445439.7 ENST00000334743.9 ENST00000557095.5 |
PPP2R5C
|
protein phosphatase 2 regulatory subunit B'gamma |
| chr18_-_11908330 | 0.16 |
ENST00000344987.11
ENST00000588103.5 ENST00000588191.5 ENST00000317235.11 ENST00000309976.13 ENST00000588186.5 ENST00000589267.5 |
MPPE1
|
metallophosphoesterase 1 |
| chr17_+_82228397 | 0.16 |
ENST00000584689.6
ENST00000392341.6 |
SLC16A3
|
solute carrier family 16 member 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.4 | 1.3 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.3 | 1.3 | GO:1903296 | regulation of glutamate secretion, neurotransmission(GO:1903294) positive regulation of glutamate secretion, neurotransmission(GO:1903296) |
| 0.3 | 0.9 | GO:1902214 | negative regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034125) regulation of interleukin-4-mediated signaling pathway(GO:1902214) |
| 0.2 | 0.7 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.2 | 0.7 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.2 | 0.7 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.2 | 0.7 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.2 | 1.3 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.2 | 1.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.2 | 0.6 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.2 | 0.5 | GO:0018315 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.2 | 0.7 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.1 | 0.1 | GO:1902567 | negative regulation of eosinophil activation(GO:1902567) |
| 0.1 | 0.5 | GO:1904045 | cellular response to aldosterone(GO:1904045) |
| 0.1 | 0.6 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 1.4 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.5 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 | 0.3 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.5 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.1 | 0.9 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.1 | 0.5 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 0.5 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.5 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.1 | 1.3 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.1 | 0.1 | GO:0001810 | regulation of type I hypersensitivity(GO:0001810) type I hypersensitivity(GO:0016068) |
| 0.1 | 0.3 | GO:0039507 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.1 | 0.5 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 0.5 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 1.3 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.1 | 0.3 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 1.8 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.1 | 0.4 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 0.3 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 0.3 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.1 | 0.3 | GO:2000612 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 | 1.4 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.1 | 0.2 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 0.2 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.1 | 0.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.4 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.1 | 0.2 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 | 0.2 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 0.2 | GO:0021592 | midbrain-hindbrain boundary morphogenesis(GO:0021555) fourth ventricle development(GO:0021592) |
| 0.1 | 0.3 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 0.1 | 1.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.6 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.3 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) secretory granule localization(GO:0032252) |
| 0.0 | 0.1 | GO:0002238 | response to molecule of fungal origin(GO:0002238) cellular response to molecule of fungal origin(GO:0071226) |
| 0.0 | 0.4 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.2 | GO:0001923 | B-1 B cell differentiation(GO:0001923) B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.2 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.3 | GO:0001767 | establishment of lymphocyte polarity(GO:0001767) |
| 0.0 | 1.1 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.4 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.5 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.2 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.2 | GO:0034239 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.0 | 0.9 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.3 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.5 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 0.0 | 0.3 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 1.1 | GO:0051412 | response to corticosterone(GO:0051412) |
| 0.0 | 0.1 | GO:0002290 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.0 | 0.3 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.1 | GO:1902336 | vestibulocochlear nerve structural organization(GO:0021649) neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) positive regulation of retinal ganglion cell axon guidance(GO:1902336) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.0 | 0.1 | GO:0071656 | negative regulation of interleukin-3 production(GO:0032712) negative regulation of granulocyte colony-stimulating factor production(GO:0071656) negative regulation of macrophage colony-stimulating factor production(GO:1901257) |
| 0.0 | 0.1 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.2 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.2 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.2 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.3 | GO:0072672 | neutrophil extravasation(GO:0072672) |
| 0.0 | 0.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.6 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 1.7 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.4 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.0 | 0.1 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.1 | GO:0002194 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.0 | 0.3 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.3 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.2 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.0 | 0.2 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.1 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.0 | 0.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.4 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.8 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.3 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.2 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 | 0.1 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.1 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.8 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 0.6 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.2 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.4 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.3 | GO:0015669 | gas transport(GO:0015669) oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.0 | 0.4 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.2 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.3 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.2 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.1 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.2 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.8 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 1.2 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.2 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.2 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0032449 | CBM complex(GO:0032449) |
| 0.2 | 1.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 0.5 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 1.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.6 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.9 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 0.4 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 0.3 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.1 | 0.4 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.7 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.1 | 0.9 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.5 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.3 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 1.2 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.3 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.2 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.3 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 0.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.1 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.0 | 0.3 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 1.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.0 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.3 | 1.9 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.3 | 1.3 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.2 | 0.9 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.2 | 0.7 | GO:0046921 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.2 | 0.5 | GO:0008940 | nitrate reductase activity(GO:0008940) molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.2 | 1.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.4 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.1 | 0.4 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.1 | 1.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.9 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.1 | 0.4 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 1.0 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.5 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 0.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.2 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.1 | 0.6 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.3 | GO:1904492 | Ac-Asp-Glu binding(GO:1904492) tetrahydrofolyl-poly(glutamate) polymer binding(GO:1904493) |
| 0.1 | 0.3 | GO:0001626 | nociceptin receptor activity(GO:0001626) |
| 0.1 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.5 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.7 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 0.2 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 0.1 | 1.6 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.5 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 1.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.4 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.9 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.5 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.3 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.4 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.5 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.1 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 1.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.4 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.3 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.3 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.4 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 1.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.3 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.6 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.3 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 1.4 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.3 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.7 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.5 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.0 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0016849 | phosphorus-oxygen lyase activity(GO:0016849) |
| 0.0 | 1.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0032357 | oxidized purine DNA binding(GO:0032357) |
| 0.0 | 0.3 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.7 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.6 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.7 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.6 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.1 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 1.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.4 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 1.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.9 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.7 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |