Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
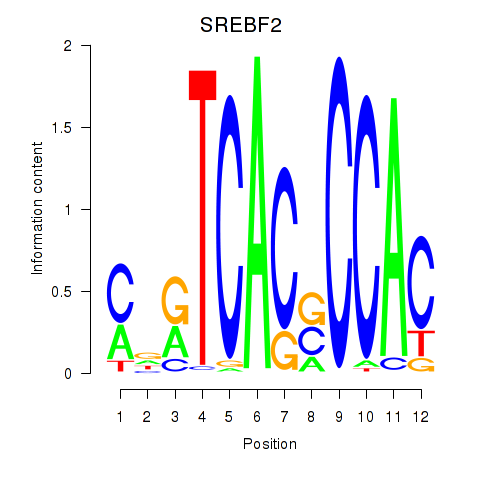
Results for SREBF2
Z-value: 0.67
Transcription factors associated with SREBF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SREBF2
|
ENSG00000198911.12 | SREBF2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SREBF2 | hg38_v1_chr22_+_41833079_41833142 | 0.62 | 9.0e-04 | Click! |
Activity profile of SREBF2 motif
Sorted Z-values of SREBF2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SREBF2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_11089446 | 2.28 |
ENST00000557933.5
ENST00000455727.6 ENST00000535915.5 ENST00000545707.5 ENST00000558518.6 ENST00000558013.5 |
LDLR
|
low density lipoprotein receptor |
| chr10_-_114684457 | 1.59 |
ENST00000392955.7
|
ABLIM1
|
actin binding LIM protein 1 |
| chr20_+_34876603 | 1.56 |
ENST00000360596.7
ENST00000253382.5 |
ACSS2
|
acyl-CoA synthetase short chain family member 2 |
| chr8_+_11802667 | 1.45 |
ENST00000443614.6
ENST00000220584.9 ENST00000525900.5 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr10_-_114684612 | 1.44 |
ENST00000533213.6
ENST00000369252.8 |
ABLIM1
|
actin binding LIM protein 1 |
| chr8_+_11802611 | 1.41 |
ENST00000623368.3
|
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr5_+_160229499 | 1.26 |
ENST00000402432.4
|
FABP6
|
fatty acid binding protein 6 |
| chr8_-_17722217 | 1.25 |
ENST00000381861.7
|
MTUS1
|
microtubule associated scaffold protein 1 |
| chr5_-_43313473 | 1.14 |
ENST00000433297.2
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 |
| chr5_-_43313403 | 1.10 |
ENST00000325110.11
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 |
| chr5_-_39424966 | 0.95 |
ENST00000515700.5
ENST00000320816.11 ENST00000339788.10 |
DAB2
|
DAB adaptor protein 2 |
| chr15_+_47184076 | 0.92 |
ENST00000558014.5
|
SEMA6D
|
semaphorin 6D |
| chr11_-_119340816 | 0.86 |
ENST00000528368.3
|
C1QTNF5
|
C1q and TNF related 5 |
| chr17_-_82098187 | 0.85 |
ENST00000634990.1
|
FASN
|
fatty acid synthase |
| chr6_-_52995170 | 0.85 |
ENST00000370959.1
ENST00000370963.9 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr18_+_79395942 | 0.83 |
ENST00000397790.6
|
NFATC1
|
nuclear factor of activated T cells 1 |
| chr15_-_72783611 | 0.80 |
ENST00000563907.5
|
ADPGK
|
ADP dependent glucokinase |
| chr10_-_50623897 | 0.75 |
ENST00000361781.7
ENST00000429490.5 ENST00000619438.4 |
SGMS1
|
sphingomyelin synthase 1 |
| chr17_+_81103998 | 0.72 |
ENST00000572498.1
|
BAIAP2
|
BAR/IMD domain containing adaptor protein 2 |
| chr8_+_123416766 | 0.65 |
ENST00000287387.7
ENST00000650311.1 ENST00000523356.1 |
NTAQ1
|
N-terminal glutamine amidase 1 |
| chr8_+_123416718 | 0.64 |
ENST00000523984.5
|
NTAQ1
|
N-terminal glutamine amidase 1 |
| chr2_-_55923775 | 0.62 |
ENST00000438672.5
ENST00000355426.8 ENST00000440439.5 ENST00000429909.5 ENST00000424207.5 ENST00000452337.5 ENST00000439193.5 ENST00000421664.1 |
EFEMP1
|
EGF containing fibulin extracellular matrix protein 1 |
| chr6_-_32184227 | 0.59 |
ENST00000450110.5
ENST00000375067.7 ENST00000375056.6 |
AGER
|
advanced glycosylation end-product specific receptor |
| chr17_-_41918966 | 0.54 |
ENST00000537919.5
ENST00000353196.5 ENST00000393896.6 |
ACLY
|
ATP citrate lyase |
| chr2_-_25982471 | 0.54 |
ENST00000264712.8
|
KIF3C
|
kinesin family member 3C |
| chr5_-_150155828 | 0.53 |
ENST00000261799.9
|
PDGFRB
|
platelet derived growth factor receptor beta |
| chr10_-_3785179 | 0.51 |
ENST00000469435.1
|
KLF6
|
Kruppel like factor 6 |
| chr19_-_18281612 | 0.50 |
ENST00000252818.5
|
JUND
|
JunD proto-oncogene, AP-1 transcription factor subunit |
| chr8_+_123416735 | 0.49 |
ENST00000524254.5
|
NTAQ1
|
N-terminal glutamine amidase 1 |
| chr22_+_41833079 | 0.48 |
ENST00000612482.4
ENST00000361204.9 |
SREBF2
|
sterol regulatory element binding transcription factor 2 |
| chr5_-_108367860 | 0.48 |
ENST00000496714.2
|
FBXL17
|
F-box and leucine rich repeat protein 17 |
| chr11_-_73598067 | 0.47 |
ENST00000450446.6
ENST00000356467.5 |
FAM168A
|
family with sequence similarity 168 member A |
| chr11_-_6655788 | 0.46 |
ENST00000299441.5
|
DCHS1
|
dachsous cadherin-related 1 |
| chr6_-_32184287 | 0.42 |
ENST00000375069.7
ENST00000375070.7 ENST00000438221.6 ENST00000620802.4 ENST00000538695.2 |
AGER
|
advanced glycosylation end-product specific receptor |
| chr17_-_41918944 | 0.42 |
ENST00000352035.7
ENST00000590770.5 ENST00000590151.5 |
ACLY
|
ATP citrate lyase |
| chr11_-_73598183 | 0.41 |
ENST00000064778.8
|
FAM168A
|
family with sequence similarity 168 member A |
| chr15_-_72783685 | 0.40 |
ENST00000456471.3
ENST00000311669.12 |
ADPGK
|
ADP dependent glucokinase |
| chr6_-_32184243 | 0.39 |
ENST00000375055.6
ENST00000375076.9 |
AGER
|
advanced glycosylation end-product specific receptor |
| chr8_-_123416327 | 0.38 |
ENST00000521903.5
|
ATAD2
|
ATPase family AAA domain containing 2 |
| chr12_-_48004467 | 0.35 |
ENST00000380518.8
|
COL2A1
|
collagen type II alpha 1 chain |
| chr10_-_3785197 | 0.34 |
ENST00000497571.6
|
KLF6
|
Kruppel like factor 6 |
| chr16_+_2033264 | 0.34 |
ENST00000565855.5
ENST00000566198.1 |
SLC9A3R2
|
SLC9A3 regulator 2 |
| chr3_-_56683218 | 0.31 |
ENST00000355628.9
ENST00000683822.1 |
TASOR
|
transcription activation suppressor |
| chr10_-_3785225 | 0.31 |
ENST00000542957.1
|
KLF6
|
Kruppel like factor 6 |
| chr12_-_48004496 | 0.29 |
ENST00000337299.7
|
COL2A1
|
collagen type II alpha 1 chain |
| chr16_+_66880503 | 0.29 |
ENST00000568869.1
ENST00000311765.4 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr3_+_183253795 | 0.28 |
ENST00000460419.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr6_+_41637005 | 0.26 |
ENST00000419164.6
ENST00000373051.6 |
MDFI
|
MyoD family inhibitor |
| chrX_-_112840815 | 0.26 |
ENST00000304758.5
ENST00000371959.9 |
AMOT
|
angiomotin |
| chr3_+_50269140 | 0.24 |
ENST00000616701.5
ENST00000433753.4 ENST00000611067.4 |
SEMA3B
|
semaphorin 3B |
| chr6_+_78867524 | 0.24 |
ENST00000369940.7
|
IRAK1BP1
|
interleukin 1 receptor associated kinase 1 binding protein 1 |
| chr11_+_64291992 | 0.23 |
ENST00000394525.6
|
KCNK4
|
potassium two pore domain channel subfamily K member 4 |
| chr8_+_26577843 | 0.23 |
ENST00000311151.9
|
DPYSL2
|
dihydropyrimidinase like 2 |
| chr6_+_151239951 | 0.22 |
ENST00000402676.7
|
AKAP12
|
A-kinase anchoring protein 12 |
| chr3_-_160399512 | 0.21 |
ENST00000498409.5
ENST00000475677.5 ENST00000478536.1 |
IFT80
|
intraflagellar transport 80 |
| chr16_+_31214088 | 0.21 |
ENST00000613872.1
|
TRIM72
|
tripartite motif containing 72 |
| chr16_+_31214111 | 0.20 |
ENST00000322122.8
|
TRIM72
|
tripartite motif containing 72 |
| chr2_+_119759875 | 0.18 |
ENST00000263708.7
|
PTPN4
|
protein tyrosine phosphatase non-receptor type 4 |
| chr15_+_78264552 | 0.16 |
ENST00000394852.8
ENST00000343789.7 |
DNAJA4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr12_-_57846686 | 0.16 |
ENST00000548823.1
ENST00000398073.7 |
CTDSP2
|
CTD small phosphatase 2 |
| chr1_+_55039511 | 0.14 |
ENST00000302118.5
|
PCSK9
|
proprotein convertase subtilisin/kexin type 9 |
| chr1_+_32753991 | 0.12 |
ENST00000373481.7
|
KIAA1522
|
KIAA1522 |
| chr14_-_74493275 | 0.10 |
ENST00000541064.5
|
NPC2
|
NPC intracellular cholesterol transporter 2 |
| chr19_+_55080363 | 0.09 |
ENST00000588359.5
ENST00000245618.5 |
EPS8L1
|
EPS8 like 1 |
| chr14_-_74493291 | 0.09 |
ENST00000238633.6
ENST00000555619.6 ENST00000434013.6 |
NPC2
|
NPC intracellular cholesterol transporter 2 |
| chr14_-_74493322 | 0.08 |
ENST00000553490.5
ENST00000557510.5 |
NPC2
|
NPC intracellular cholesterol transporter 2 |
| chr11_+_22666604 | 0.08 |
ENST00000454584.6
|
GAS2
|
growth arrest specific 2 |
| chr11_+_2902258 | 0.06 |
ENST00000649076.2
ENST00000449793.6 |
SLC22A18
|
solute carrier family 22 member 18 |
| chr9_+_128689201 | 0.06 |
ENST00000322030.13
|
SET
|
SET nuclear proto-oncogene |
| chr11_+_2902388 | 0.06 |
ENST00000380574.5
|
SLC22A18
|
solute carrier family 22 member 18 |
| chr1_+_197268222 | 0.06 |
ENST00000367400.8
ENST00000638467.1 ENST00000367399.6 |
CRB1
|
crumbs cell polarity complex component 1 |
| chr17_-_41118369 | 0.05 |
ENST00000391413.4
|
KRTAP4-11
|
keratin associated protein 4-11 |
| chr12_+_49950733 | 0.05 |
ENST00000199280.4
ENST00000550862.1 |
AQP2
|
aquaporin 2 |
| chr2_-_219060914 | 0.04 |
ENST00000295731.7
|
IHH
|
Indian hedgehog signaling molecule |
| chr18_-_23586422 | 0.02 |
ENST00000269228.10
|
NPC1
|
NPC intracellular cholesterol transporter 1 |
| chr2_+_231707650 | 0.02 |
ENST00000409321.5
|
PTMA
|
prothymosin alpha |
| chr9_+_128689948 | 0.02 |
ENST00000372688.8
ENST00000372686.5 |
SET
|
SET nuclear proto-oncogene |
| chr19_-_13938371 | 0.02 |
ENST00000588872.3
ENST00000339560.10 |
PODNL1
|
podocan like 1 |
| chr19_-_45782388 | 0.01 |
ENST00000458663.6
|
DMPK
|
DM1 protein kinase |
| chr11_+_65711991 | 0.01 |
ENST00000377046.7
ENST00000352980.8 |
KAT5
|
lysine acetyltransferase 5 |
| chr1_+_197268204 | 0.01 |
ENST00000535699.5
ENST00000538660.5 |
CRB1
|
crumbs cell polarity complex component 1 |
| chr3_+_160399630 | 0.00 |
ENST00000465903.5
ENST00000485645.5 ENST00000472991.5 ENST00000467468.5 ENST00000469762.5 ENST00000357388.8 ENST00000489573.5 ENST00000462787.5 ENST00000490207.5 ENST00000485867.5 |
SMC4
|
structural maintenance of chromosomes 4 |
| chr19_-_11197516 | 0.00 |
ENST00000592903.5
ENST00000586659.6 ENST00000589359.5 ENST00000588724.5 |
KANK2
|
KN motif and ankyrin repeat domains 2 |
| chr11_+_65615755 | 0.00 |
ENST00000355703.4
|
PCNX3
|
pecanex 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.5 | 1.6 | GO:0019542 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.5 | 2.9 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.5 | 1.4 | GO:1904597 | regulation of connective tissue replacement involved in inflammatory response wound healing(GO:1904596) negative regulation of connective tissue replacement involved in inflammatory response wound healing(GO:1904597) regulation of advanced glycation end-product receptor activity(GO:1904603) negative regulation of advanced glycation end-product receptor activity(GO:1904604) negative regulation of connective tissue replacement(GO:1905204) |
| 0.3 | 1.0 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.3 | 0.9 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.2 | 0.5 | GO:0072275 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.2 | 0.5 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.1 | 2.2 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 | 0.5 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 0.6 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.1 | 0.8 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.7 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.1 | 0.2 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.1 | 0.9 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 1.0 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.1 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.3 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.6 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.0 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.4 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.4 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 1.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.8 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.9 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.0 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 1.2 | GO:0006735 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 2.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.3 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.3 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 0.3 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.8 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.8 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.9 | GO:0070206 | protein trimerization(GO:0070206) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.2 | 1.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.5 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.9 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.8 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.5 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.5 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 3.0 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.8 | GO:0097542 | ciliary tip(GO:0097542) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.5 | 2.9 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.5 | 1.4 | GO:1904599 | advanced glycation end-product binding(GO:1904599) |
| 0.4 | 1.6 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.3 | 2.3 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.3 | 0.9 | GO:0016420 | [acyl-carrier-protein] S-malonyltransferase activity(GO:0004314) 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) S-malonyltransferase activity(GO:0016419) malonyltransferase activity(GO:0016420) |
| 0.2 | 1.0 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.2 | 0.8 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 0.5 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.2 | 0.5 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.2 | 0.6 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.1 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.1 | 0.8 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.1 | 0.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.2 | GO:0097604 | temperature-gated cation channel activity(GO:0097604) |
| 0.1 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 1.3 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.3 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.2 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 0.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.0 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 2.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.5 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.8 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 2.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 3.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 0.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 1.0 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.9 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |