Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for SRY
Z-value: 0.93
Transcription factors associated with SRY
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SRY
|
ENSG00000184895.8 | SRY |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SRY | hg38_v1_chrY_-_2787676_2787687 | -0.21 | 3.1e-01 | Click! |
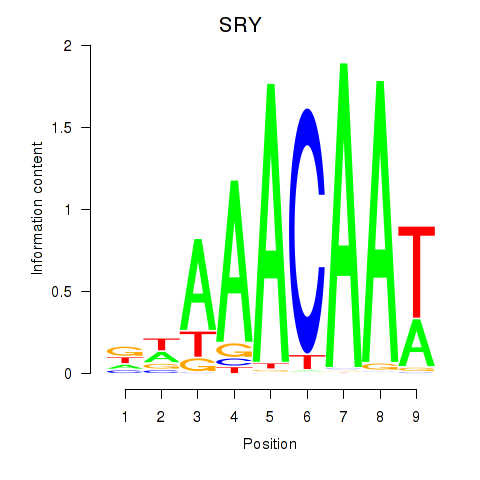
Activity profile of SRY motif
Sorted Z-values of SRY motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SRY
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_41612892 | 4.59 |
ENST00000509454.5
ENST00000396595.7 ENST00000381753.8 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr4_+_41612702 | 4.26 |
ENST00000509277.5
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr5_-_111756245 | 3.55 |
ENST00000447165.6
|
NREP
|
neuronal regeneration related protein |
| chr9_-_14314132 | 2.89 |
ENST00000380953.6
|
NFIB
|
nuclear factor I B |
| chr9_-_14314567 | 2.55 |
ENST00000397579.6
|
NFIB
|
nuclear factor I B |
| chr9_-_14314519 | 2.45 |
ENST00000397581.6
|
NFIB
|
nuclear factor I B |
| chr1_+_61081728 | 2.39 |
ENST00000371189.8
|
NFIA
|
nuclear factor I A |
| chr1_+_84164370 | 2.36 |
ENST00000446538.5
ENST00000610703.4 ENST00000370682.7 ENST00000394838.6 ENST00000432111.5 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr1_+_84164962 | 2.24 |
ENST00000614872.4
ENST00000394839.6 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr2_-_156342348 | 2.22 |
ENST00000409572.5
|
NR4A2
|
nuclear receptor subfamily 4 group A member 2 |
| chr14_-_91946989 | 2.21 |
ENST00000556154.5
|
FBLN5
|
fibulin 5 |
| chr14_+_52553273 | 2.09 |
ENST00000542169.6
ENST00000555622.1 |
GPR137C
|
G protein-coupled receptor 137C |
| chr17_-_69141878 | 1.98 |
ENST00000590645.1
ENST00000284425.7 |
ABCA6
|
ATP binding cassette subfamily A member 6 |
| chr1_+_61082553 | 1.90 |
ENST00000403491.8
ENST00000371187.7 |
NFIA
|
nuclear factor I A |
| chr14_-_91947654 | 1.90 |
ENST00000342058.9
|
FBLN5
|
fibulin 5 |
| chr18_+_23135452 | 1.78 |
ENST00000580153.5
ENST00000256925.12 |
CABLES1
|
Cdk5 and Abl enzyme substrate 1 |
| chr9_-_14313642 | 1.72 |
ENST00000637742.1
|
NFIB
|
nuclear factor I B |
| chr9_+_2159672 | 1.71 |
ENST00000634343.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr15_-_59372863 | 1.64 |
ENST00000288235.9
|
MYO1E
|
myosin IE |
| chr14_-_91947383 | 1.64 |
ENST00000267620.14
|
FBLN5
|
fibulin 5 |
| chr17_-_64390852 | 1.62 |
ENST00000563924.6
|
PECAM1
|
platelet and endothelial cell adhesion molecule 1 |
| chr18_+_75210789 | 1.47 |
ENST00000580243.3
|
TSHZ1
|
teashirt zinc finger homeobox 1 |
| chr18_-_55635948 | 1.44 |
ENST00000565124.4
ENST00000398339.5 |
TCF4
|
transcription factor 4 |
| chr1_+_164559739 | 1.43 |
ENST00000627490.2
|
PBX1
|
PBX homeobox 1 |
| chr9_-_127847117 | 1.42 |
ENST00000480266.5
|
ENG
|
endoglin |
| chr18_+_75210755 | 1.41 |
ENST00000322038.5
|
TSHZ1
|
teashirt zinc finger homeobox 1 |
| chr9_+_2110354 | 1.35 |
ENST00000634772.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr1_+_185734362 | 1.32 |
ENST00000271588.9
|
HMCN1
|
hemicentin 1 |
| chr15_-_70702273 | 1.31 |
ENST00000558758.5
ENST00000379983.6 ENST00000560441.5 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr16_+_30395400 | 1.25 |
ENST00000320159.2
ENST00000613509.2 |
ZNF48
|
zinc finger protein 48 |
| chr2_-_208254232 | 1.19 |
ENST00000415913.5
ENST00000415282.5 ENST00000446179.5 |
IDH1
|
isocitrate dehydrogenase (NADP(+)) 1 |
| chr14_-_22819721 | 1.17 |
ENST00000554517.5
ENST00000285850.11 ENST00000397529.6 ENST00000555702.5 |
SLC7A7
|
solute carrier family 7 member 7 |
| chr7_-_32892015 | 1.10 |
ENST00000452926.1
|
KBTBD2
|
kelch repeat and BTB domain containing 2 |
| chr18_+_3448456 | 1.08 |
ENST00000549780.5
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr9_-_14313843 | 1.06 |
ENST00000636063.1
ENST00000380921.3 ENST00000622520.1 ENST00000380959.7 |
NFIB
|
nuclear factor I B |
| chr14_+_52552830 | 1.03 |
ENST00000321662.11
|
GPR137C
|
G protein-coupled receptor 137C |
| chr14_-_89417148 | 0.98 |
ENST00000557258.6
|
FOXN3
|
forkhead box N3 |
| chrX_+_28587411 | 0.96 |
ENST00000378993.6
|
IL1RAPL1
|
interleukin 1 receptor accessory protein like 1 |
| chr1_+_33256479 | 0.94 |
ENST00000539719.6
ENST00000483388.5 |
ZNF362
|
zinc finger protein 362 |
| chr18_-_55302613 | 0.93 |
ENST00000561831.7
|
TCF4
|
transcription factor 4 |
| chr3_-_142149515 | 0.93 |
ENST00000475734.5
ENST00000467072.5 ENST00000489671.6 |
TFDP2
|
transcription factor Dp-2 |
| chr18_-_55589836 | 0.88 |
ENST00000537578.5
ENST00000564403.6 |
TCF4
|
transcription factor 4 |
| chr18_-_55589770 | 0.87 |
ENST00000565018.6
ENST00000636400.2 |
TCF4
|
transcription factor 4 |
| chr18_-_55321986 | 0.86 |
ENST00000570287.6
|
TCF4
|
transcription factor 4 |
| chr14_+_91114431 | 0.85 |
ENST00000428926.6
ENST00000517362.5 |
DGLUCY
|
D-glutamate cyclase |
| chrX_+_85244075 | 0.84 |
ENST00000276123.7
|
ZNF711
|
zinc finger protein 711 |
| chrX_+_85244032 | 0.83 |
ENST00000373165.7
|
ZNF711
|
zinc finger protein 711 |
| chr14_+_91114667 | 0.81 |
ENST00000523894.5
ENST00000522322.5 ENST00000523771.5 |
DGLUCY
|
D-glutamate cyclase |
| chr18_-_55321640 | 0.80 |
ENST00000637169.2
|
TCF4
|
transcription factor 4 |
| chr18_-_55589795 | 0.80 |
ENST00000568740.5
ENST00000629387.2 |
TCF4
|
transcription factor 4 |
| chr3_+_28348695 | 0.79 |
ENST00000383768.7
|
ZCWPW2
|
zinc finger CW-type and PWWP domain containing 2 |
| chr17_+_59331633 | 0.77 |
ENST00000312655.9
|
YPEL2
|
yippee like 2 |
| chr4_-_151325488 | 0.77 |
ENST00000604030.7
|
SH3D19
|
SH3 domain containing 19 |
| chr14_+_91114364 | 0.76 |
ENST00000518868.5
|
DGLUCY
|
D-glutamate cyclase |
| chr12_-_102197827 | 0.76 |
ENST00000329406.5
|
PMCH
|
pro-melanin concentrating hormone |
| chr2_-_159616442 | 0.75 |
ENST00000541068.6
ENST00000392783.7 ENST00000392782.5 |
BAZ2B
|
bromodomain adjacent to zinc finger domain 2B |
| chr14_-_52552493 | 0.74 |
ENST00000281741.9
ENST00000557374.1 |
TXNDC16
|
thioredoxin domain containing 16 |
| chr1_+_96722628 | 0.74 |
ENST00000675401.1
|
PTBP2
|
polypyrimidine tract binding protein 2 |
| chr2_-_191847068 | 0.74 |
ENST00000304141.5
|
CAVIN2
|
caveolae associated protein 2 |
| chr3_-_28348805 | 0.73 |
ENST00000457172.5
ENST00000479665.6 |
AZI2
|
5-azacytidine induced 2 |
| chr18_-_55322215 | 0.72 |
ENST00000457482.7
|
TCF4
|
transcription factor 4 |
| chr21_-_38498415 | 0.72 |
ENST00000398905.5
ENST00000398907.5 ENST00000453032.6 ENST00000288319.12 |
ERG
|
ETS transcription factor ERG |
| chr9_-_71768386 | 0.70 |
ENST00000377066.9
ENST00000377044.9 |
CEMIP2
|
cell migration inducing hyaluronidase 2 |
| chr3_-_112641292 | 0.70 |
ENST00000439685.6
|
CCDC80
|
coiled-coil domain containing 80 |
| chr2_+_108588286 | 0.69 |
ENST00000332345.10
|
LIMS1
|
LIM zinc finger domain containing 1 |
| chr13_-_74133892 | 0.68 |
ENST00000377669.7
|
KLF12
|
Kruppel like factor 12 |
| chr16_-_15643024 | 0.67 |
ENST00000540441.6
|
MARF1
|
meiosis regulator and mRNA stability factor 1 |
| chr1_+_84164684 | 0.67 |
ENST00000370680.5
|
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr6_+_156777366 | 0.66 |
ENST00000636930.2
|
ARID1B
|
AT-rich interaction domain 1B |
| chr1_+_66332004 | 0.66 |
ENST00000371045.9
ENST00000531025.5 ENST00000526197.5 |
PDE4B
|
phosphodiesterase 4B |
| chr19_+_10602436 | 0.65 |
ENST00000590382.5
ENST00000407327.8 |
SLC44A2
|
solute carrier family 44 member 2 |
| chr14_-_50561119 | 0.60 |
ENST00000555216.5
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr11_-_10808304 | 0.58 |
ENST00000532082.6
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma 2 |
| chr2_+_74002685 | 0.58 |
ENST00000305799.8
|
TET3
|
tet methylcytosine dioxygenase 3 |
| chr22_+_19723525 | 0.58 |
ENST00000366425.4
|
GP1BB
|
glycoprotein Ib platelet subunit beta |
| chr20_-_63951122 | 0.57 |
ENST00000369908.9
|
UCKL1
|
uridine-cytidine kinase 1 like 1 |
| chr11_+_114060204 | 0.56 |
ENST00000683318.1
|
ZBTB16
|
zinc finger and BTB domain containing 16 |
| chr5_+_139341875 | 0.56 |
ENST00000511706.5
|
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr19_+_13023958 | 0.56 |
ENST00000587760.5
ENST00000585575.5 |
NFIX
|
nuclear factor I X |
| chr12_-_85836372 | 0.56 |
ENST00000361228.5
|
RASSF9
|
Ras association domain family member 9 |
| chr10_-_14572123 | 0.54 |
ENST00000378465.7
ENST00000452706.6 ENST00000622567.4 ENST00000378458.6 |
FAM107B
|
family with sequence similarity 107 member B |
| chr6_+_138773747 | 0.54 |
ENST00000617445.5
|
CCDC28A
|
coiled-coil domain containing 28A |
| chr4_+_169660062 | 0.52 |
ENST00000507875.5
ENST00000613795.4 |
CLCN3
|
chloride voltage-gated channel 3 |
| chr3_-_28348924 | 0.51 |
ENST00000414162.5
ENST00000420543.6 |
AZI2
|
5-azacytidine induced 2 |
| chr8_+_69492793 | 0.50 |
ENST00000616868.1
ENST00000419716.7 ENST00000402687.9 |
SULF1
|
sulfatase 1 |
| chr3_+_25428233 | 0.49 |
ENST00000437042.6
ENST00000330688.9 |
RARB
|
retinoic acid receptor beta |
| chr18_-_55351977 | 0.49 |
ENST00000643689.1
|
TCF4
|
transcription factor 4 |
| chr7_-_32891744 | 0.49 |
ENST00000304056.9
|
KBTBD2
|
kelch repeat and BTB domain containing 2 |
| chr9_+_27109393 | 0.48 |
ENST00000406359.8
|
TEK
|
TEK receptor tyrosine kinase |
| chr20_+_325536 | 0.48 |
ENST00000342665.5
|
SOX12
|
SRY-box transcription factor 12 |
| chr3_+_124384513 | 0.47 |
ENST00000682540.1
ENST00000522553.6 ENST00000682695.1 ENST00000682674.1 ENST00000684382.1 |
KALRN
|
kalirin RhoGEF kinase |
| chr9_-_122227525 | 0.47 |
ENST00000373755.6
ENST00000373754.6 |
LHX6
|
LIM homeobox 6 |
| chr8_-_81483226 | 0.46 |
ENST00000256104.5
|
FABP4
|
fatty acid binding protein 4 |
| chr15_-_34318761 | 0.45 |
ENST00000290209.9
|
SLC12A6
|
solute carrier family 12 member 6 |
| chr5_+_139308095 | 0.44 |
ENST00000515833.2
|
MATR3
|
matrin 3 |
| chr15_-_70097874 | 0.43 |
ENST00000557997.5
ENST00000317509.12 ENST00000451782.7 ENST00000627388.2 |
TLE3
|
TLE family member 3, transcriptional corepressor |
| chr11_-_82997477 | 0.42 |
ENST00000534301.5
|
RAB30
|
RAB30, member RAS oncogene family |
| chr11_+_128694052 | 0.42 |
ENST00000527786.7
ENST00000534087.3 |
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr18_+_3451647 | 0.42 |
ENST00000345133.9
ENST00000330513.10 ENST00000549546.5 |
TGIF1
|
TGFB induced factor homeobox 1 |
| chr12_+_59664677 | 0.41 |
ENST00000548610.5
|
SLC16A7
|
solute carrier family 16 member 7 |
| chr2_+_108588453 | 0.40 |
ENST00000393310.5
|
LIMS1
|
LIM zinc finger domain containing 1 |
| chr19_+_13024917 | 0.39 |
ENST00000587260.1
|
NFIX
|
nuclear factor I X |
| chrX_-_19670983 | 0.39 |
ENST00000379716.5
|
SH3KBP1
|
SH3 domain containing kinase binding protein 1 |
| chr18_+_3451585 | 0.38 |
ENST00000551541.5
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr7_-_27180230 | 0.38 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr3_-_28348629 | 0.38 |
ENST00000334100.10
|
AZI2
|
5-azacytidine induced 2 |
| chr11_+_128693887 | 0.37 |
ENST00000281428.12
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr16_-_1782526 | 0.37 |
ENST00000566339.6
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chr7_+_80638633 | 0.37 |
ENST00000447544.7
ENST00000482059.6 |
CD36
|
CD36 molecule |
| chrX_+_123961696 | 0.35 |
ENST00000371145.8
ENST00000371157.7 ENST00000371144.7 |
STAG2
|
stromal antigen 2 |
| chr3_+_189789672 | 0.35 |
ENST00000434928.5
|
TP63
|
tumor protein p63 |
| chr2_+_69013379 | 0.35 |
ENST00000409349.7
|
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr14_+_22516273 | 0.35 |
ENST00000390510.1
|
TRAJ27
|
T cell receptor alpha joining 27 |
| chr20_-_44521989 | 0.35 |
ENST00000342374.5
ENST00000255175.5 |
SERINC3
|
serine incorporator 3 |
| chr10_+_113125536 | 0.34 |
ENST00000349937.7
|
TCF7L2
|
transcription factor 7 like 2 |
| chr5_-_11589019 | 0.34 |
ENST00000511377.5
|
CTNND2
|
catenin delta 2 |
| chr19_-_45768627 | 0.34 |
ENST00000560160.1
|
SIX5
|
SIX homeobox 5 |
| chrX_+_15507302 | 0.34 |
ENST00000342014.6
|
BMX
|
BMX non-receptor tyrosine kinase |
| chr1_-_52552994 | 0.33 |
ENST00000355809.4
ENST00000528642.5 ENST00000470626.1 ENST00000257177.9 ENST00000371544.7 |
TUT4
|
terminal uridylyl transferase 4 |
| chr7_+_80638510 | 0.33 |
ENST00000433696.6
ENST00000538969.5 ENST00000544133.5 |
CD36
|
CD36 molecule |
| chr6_+_156777882 | 0.33 |
ENST00000350026.10
ENST00000647938.1 ENST00000674298.1 |
ARID1B
|
AT-rich interaction domain 1B |
| chr13_-_40666600 | 0.33 |
ENST00000379561.6
|
FOXO1
|
forkhead box O1 |
| chr5_+_139342442 | 0.33 |
ENST00000394795.6
ENST00000510080.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr14_-_73027077 | 0.32 |
ENST00000553891.5
ENST00000556143.6 |
ZFYVE1
|
zinc finger FYVE-type containing 1 |
| chr5_+_141177790 | 0.32 |
ENST00000239444.4
ENST00000623995.1 |
PCDHB8
ENSG00000279472.1
|
protocadherin beta 8 novel transcript |
| chr8_+_66493556 | 0.31 |
ENST00000305454.8
ENST00000522977.5 ENST00000480005.1 |
VXN
|
vexin |
| chr2_+_190408324 | 0.31 |
ENST00000417958.5
ENST00000432036.5 ENST00000392328.6 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chrX_-_24672654 | 0.31 |
ENST00000379145.5
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr7_+_134866831 | 0.30 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr10_-_125160499 | 0.30 |
ENST00000494626.6
ENST00000337195.9 |
CTBP2
|
C-terminal binding protein 2 |
| chr9_-_95509241 | 0.29 |
ENST00000331920.11
|
PTCH1
|
patched 1 |
| chr14_-_73027117 | 0.29 |
ENST00000318876.9
|
ZFYVE1
|
zinc finger FYVE-type containing 1 |
| chr2_-_207165923 | 0.29 |
ENST00000309446.11
|
KLF7
|
Kruppel like factor 7 |
| chr20_-_3781440 | 0.29 |
ENST00000379756.3
|
SPEF1
|
sperm flagellar 1 |
| chr3_+_179148341 | 0.29 |
ENST00000263967.4
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chrX_+_101488044 | 0.28 |
ENST00000423738.4
|
ARMCX4
|
armadillo repeat containing X-linked 4 |
| chr10_+_110207587 | 0.28 |
ENST00000332674.9
ENST00000453116.5 |
MXI1
|
MAX interactor 1, dimerization protein |
| chr18_+_11851404 | 0.28 |
ENST00000526991.3
|
CHMP1B
|
charged multivesicular body protein 1B |
| chr18_+_21363593 | 0.28 |
ENST00000580732.6
|
GREB1L
|
GREB1 like retinoic acid receptor coactivator |
| chr7_+_143288215 | 0.28 |
ENST00000619992.4
ENST00000310447.10 |
CASP2
|
caspase 2 |
| chr1_-_151146643 | 0.26 |
ENST00000613223.1
|
SEMA6C
|
semaphorin 6C |
| chr2_+_86720282 | 0.26 |
ENST00000283632.5
|
RMND5A
|
required for meiotic nuclear division 5 homolog A |
| chr17_-_42276341 | 0.25 |
ENST00000293328.8
|
STAT5B
|
signal transducer and activator of transcription 5B |
| chr1_+_206834347 | 0.25 |
ENST00000340758.7
|
IL19
|
interleukin 19 |
| chr10_+_72893734 | 0.24 |
ENST00000334011.10
|
OIT3
|
oncoprotein induced transcript 3 |
| chr5_+_87267792 | 0.24 |
ENST00000274376.11
|
RASA1
|
RAS p21 protein activator 1 |
| chr17_-_59151794 | 0.24 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr7_+_128758947 | 0.24 |
ENST00000493278.1
|
CALU
|
calumenin |
| chr11_-_118225002 | 0.24 |
ENST00000356289.10
ENST00000526620.5 |
JAML
|
junction adhesion molecule like |
| chr2_+_27442421 | 0.24 |
ENST00000407293.5
|
KRTCAP3
|
keratinocyte associated protein 3 |
| chr2_+_9961165 | 0.23 |
ENST00000405379.6
|
GRHL1
|
grainyhead like transcription factor 1 |
| chr10_+_110225955 | 0.23 |
ENST00000239007.11
|
MXI1
|
MAX interactor 1, dimerization protein |
| chr8_-_71547626 | 0.23 |
ENST00000647540.1
ENST00000644229.1 |
EYA1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr19_+_13795434 | 0.22 |
ENST00000254323.6
|
ZSWIM4
|
zinc finger SWIM-type containing 4 |
| chr16_+_28846674 | 0.22 |
ENST00000322610.12
|
SH2B1
|
SH2B adaptor protein 1 |
| chr20_+_10435283 | 0.22 |
ENST00000334534.10
|
SLX4IP
|
SLX4 interacting protein |
| chr9_+_27109135 | 0.22 |
ENST00000519097.5
ENST00000615002.4 |
TEK
|
TEK receptor tyrosine kinase |
| chr10_-_48274567 | 0.22 |
ENST00000636244.1
ENST00000374201.8 |
FRMPD2
|
FERM and PDZ domain containing 2 |
| chr15_-_51971740 | 0.22 |
ENST00000315141.5
ENST00000299601.10 |
LEO1
|
LEO1 homolog, Paf1/RNA polymerase II complex component |
| chr1_+_226063466 | 0.21 |
ENST00000666609.1
ENST00000661429.1 |
H3-3A
|
H3.3 histone A |
| chr5_+_141364153 | 0.21 |
ENST00000518069.2
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr7_-_19773569 | 0.21 |
ENST00000422233.5
ENST00000433641.5 ENST00000405844.6 |
TMEM196
|
transmembrane protein 196 |
| chr14_+_58427425 | 0.21 |
ENST00000619722.5
ENST00000423743.7 |
KIAA0586
|
KIAA0586 |
| chr6_-_31684040 | 0.20 |
ENST00000375863.7
|
LY6G5C
|
lymphocyte antigen 6 family member G5C |
| chr5_+_140868945 | 0.20 |
ENST00000398640.7
|
PCDHA11
|
protocadherin alpha 11 |
| chr1_-_151146611 | 0.20 |
ENST00000341697.7
ENST00000368914.8 |
SEMA6C
|
semaphorin 6C |
| chr5_+_138338256 | 0.20 |
ENST00000513056.5
ENST00000239906.10 ENST00000511276.1 |
FAM53C
|
family with sequence similarity 53 member C |
| chr6_-_52244500 | 0.20 |
ENST00000336123.5
|
IL17F
|
interleukin 17F |
| chr12_-_8662881 | 0.20 |
ENST00000433590.6
|
MFAP5
|
microfibril associated protein 5 |
| chrX_-_16869840 | 0.20 |
ENST00000380084.8
|
RBBP7
|
RB binding protein 7, chromatin remodeling factor |
| chr14_+_58427385 | 0.19 |
ENST00000354386.10
ENST00000619416.4 |
KIAA0586
|
KIAA0586 |
| chr12_-_27971970 | 0.19 |
ENST00000395872.5
ENST00000201015.8 |
PTHLH
|
parathyroid hormone like hormone |
| chr12_-_75209422 | 0.19 |
ENST00000393288.2
ENST00000540018.5 |
KCNC2
|
potassium voltage-gated channel subfamily C member 2 |
| chr5_+_126777112 | 0.19 |
ENST00000261366.10
ENST00000492190.5 ENST00000395354.1 |
LMNB1
|
lamin B1 |
| chr8_+_66493514 | 0.19 |
ENST00000521495.5
|
VXN
|
vexin |
| chr9_-_39239174 | 0.18 |
ENST00000358144.6
|
CNTNAP3
|
contactin associated protein family member 3 |
| chr5_+_143812161 | 0.18 |
ENST00000289448.4
|
HMHB1
|
histocompatibility minor HB-1 |
| chr5_+_141364231 | 0.17 |
ENST00000611914.1
|
PCDHGA5
|
protocadherin gamma subfamily A, 5 |
| chr16_-_3880678 | 0.17 |
ENST00000262367.10
|
CREBBP
|
CREB binding protein |
| chr9_+_27109200 | 0.16 |
ENST00000380036.10
|
TEK
|
TEK receptor tyrosine kinase |
| chr19_-_45769204 | 0.16 |
ENST00000317578.7
|
SIX5
|
SIX homeobox 5 |
| chr11_-_82997394 | 0.15 |
ENST00000525117.5
ENST00000532548.5 |
RAB30
|
RAB30, member RAS oncogene family |
| chr14_+_91114388 | 0.14 |
ENST00000519019.5
ENST00000523816.5 ENST00000517518.5 |
DGLUCY
|
D-glutamate cyclase |
| chr7_+_29479712 | 0.14 |
ENST00000412711.6
|
CHN2
|
chimerin 2 |
| chr12_-_75209701 | 0.14 |
ENST00000350228.6
ENST00000298972.5 |
KCNC2
|
potassium voltage-gated channel subfamily C member 2 |
| chr4_-_110198650 | 0.14 |
ENST00000394607.7
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr20_-_62427528 | 0.14 |
ENST00000252998.2
|
RBBP8NL
|
RBBP8 N-terminal like |
| chr4_-_110198579 | 0.14 |
ENST00000302274.8
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr12_-_7503841 | 0.13 |
ENST00000359156.8
|
CD163
|
CD163 molecule |
| chr5_+_177133005 | 0.13 |
ENST00000510954.5
ENST00000354179.8 |
NSD1
|
nuclear receptor binding SET domain protein 1 |
| chr11_-_44950151 | 0.13 |
ENST00000533940.5
ENST00000533937.1 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr2_+_161231078 | 0.12 |
ENST00000439442.1
|
TANK
|
TRAF family member associated NFKB activator |
| chr22_+_39901075 | 0.12 |
ENST00000344138.9
|
GRAP2
|
GRB2 related adaptor protein 2 |
| chrX_-_40096190 | 0.12 |
ENST00000679513.1
|
BCOR
|
BCL6 corepressor |
| chr14_+_74348440 | 0.11 |
ENST00000256362.5
|
VRTN
|
vertebrae development associated |
| chr12_-_91058016 | 0.11 |
ENST00000266719.4
|
KERA
|
keratocan |
| chr6_+_83853576 | 0.11 |
ENST00000369687.2
|
RIPPLY2
|
ripply transcriptional repressor 2 |
| chr8_-_42501224 | 0.11 |
ENST00000520262.6
ENST00000517366.1 |
SLC20A2
|
solute carrier family 20 member 2 |
| chr16_+_3654683 | 0.11 |
ENST00000246949.10
|
DNASE1
|
deoxyribonuclease 1 |
| chr7_+_143959927 | 0.11 |
ENST00000624504.1
|
OR2F1
|
olfactory receptor family 2 subfamily F member 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 10.7 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.8 | 5.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.7 | 2.2 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.5 | 2.9 | GO:0060023 | soft palate development(GO:0060023) |
| 0.5 | 5.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.5 | 1.4 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.4 | 1.6 | GO:0072011 | diapedesis(GO:0050904) glomerular endothelium development(GO:0072011) |
| 0.4 | 1.2 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.3 | 1.6 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.2 | 1.6 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.2 | 0.6 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.2 | 0.5 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.2 | 1.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 1.0 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.1 | 0.7 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 | 3.8 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 0.9 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 0.4 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 | 0.5 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.6 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.1 | 0.3 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.1 | 0.4 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.1 | 0.7 | GO:0070543 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 0.3 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 0.3 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.1 | 0.5 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 0.5 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.3 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 0.2 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.1 | 0.4 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.7 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.2 | GO:0045423 | regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.0 | 1.5 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.5 | GO:0046959 | habituation(GO:0046959) negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.3 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.3 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.2 | GO:0045588 | positive regulation of gamma-delta T cell differentiation(GO:0045588) |
| 0.0 | 0.2 | GO:0035696 | monocyte extravasation(GO:0035696) |
| 0.0 | 1.0 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.2 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.6 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.8 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.4 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.3 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.8 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 8.3 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.1 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 0.5 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 1.1 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.1 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.0 | 0.7 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.3 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 8.6 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.9 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.3 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.6 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.9 | GO:1900271 | regulation of long-term synaptic potentiation(GO:1900271) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.2 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.0 | 0.2 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 | 0.0 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.0 | 0.8 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.7 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.0 | 0.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.5 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.0 | 1.3 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.2 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.4 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.0 | GO:1902688 | regulation of NAD metabolic process(GO:1902688) |
| 0.0 | 2.0 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.7 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.5 | 1.4 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.4 | 10.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 5.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 0.6 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 3.7 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 0.3 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.6 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 0.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 2.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 7.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.6 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 1.2 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.7 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 1.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 2.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.4 | 7.8 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.3 | 5.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 1.4 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 0.4 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.5 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.6 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 2.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.7 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 1.9 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 0.6 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 1.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 1.0 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 0.3 | GO:0097108 | smoothened binding(GO:0005119) hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 3.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.3 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.6 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 16.6 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.5 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.6 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.3 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 4.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.8 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 2.0 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.6 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 15.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 5.8 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 8.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 3.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.6 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 5.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 10.7 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.2 | 5.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.6 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 7.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 1.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.9 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 1.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |