Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
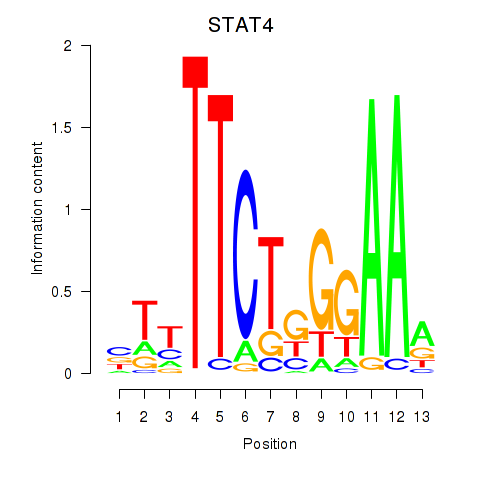
Results for STAT4
Z-value: 1.17
Transcription factors associated with STAT4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STAT4
|
ENSG00000138378.19 | STAT4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| STAT4 | hg38_v1_chr2_-_191150971_191151017, hg38_v1_chr2_-_191151568_191151596 | -0.71 | 7.3e-05 | Click! |
Activity profile of STAT4 motif
Sorted Z-values of STAT4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of STAT4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_76036060 | 9.99 |
ENST00000306621.8
|
CXCL11
|
C-X-C motif chemokine ligand 11 |
| chr6_+_32854179 | 7.42 |
ENST00000374859.3
|
PSMB9
|
proteasome 20S subunit beta 9 |
| chr15_+_67166019 | 6.79 |
ENST00000537194.6
|
SMAD3
|
SMAD family member 3 |
| chr6_-_32853618 | 6.69 |
ENST00000354258.5
|
TAP1
|
transporter 1, ATP binding cassette subfamily B member |
| chr6_-_32853813 | 6.62 |
ENST00000643049.2
|
TAP1
|
transporter 1, ATP binding cassette subfamily B member |
| chr5_+_132073782 | 6.23 |
ENST00000296871.4
|
CSF2
|
colony stimulating factor 2 |
| chr3_-_79767987 | 5.76 |
ENST00000464233.6
|
ROBO1
|
roundabout guidance receptor 1 |
| chr1_-_7940825 | 5.35 |
ENST00000377507.8
|
TNFRSF9
|
TNF receptor superfamily member 9 |
| chr17_-_35880350 | 4.99 |
ENST00000605140.6
ENST00000651122.1 ENST00000603197.6 |
CCL5
|
C-C motif chemokine ligand 5 |
| chr5_-_122078249 | 4.36 |
ENST00000231004.5
|
LOX
|
lysyl oxidase |
| chr11_+_20022550 | 4.28 |
ENST00000533917.5
|
NAV2
|
neuron navigator 2 |
| chr1_-_120100688 | 4.24 |
ENST00000652264.1
|
NOTCH2
|
notch receptor 2 |
| chr3_+_122680802 | 4.11 |
ENST00000474629.7
|
PARP14
|
poly(ADP-ribose) polymerase family member 14 |
| chr19_+_10086787 | 3.87 |
ENST00000590378.5
ENST00000397881.7 |
SHFL
|
shiftless antiviral inhibitor of ribosomal frameshifting |
| chr7_-_116030735 | 3.09 |
ENST00000393485.5
|
TFEC
|
transcription factor EC |
| chr19_+_10086305 | 2.80 |
ENST00000253110.16
ENST00000591813.5 |
SHFL
|
shiftless antiviral inhibitor of ribosomal frameshifting |
| chrX_-_155334580 | 2.73 |
ENST00000369449.7
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr2_-_201698040 | 2.60 |
ENST00000396886.7
ENST00000409143.5 |
MPP4
|
membrane palmitoylated protein 4 |
| chr14_+_24161257 | 2.60 |
ENST00000396864.8
ENST00000557894.5 ENST00000559284.5 ENST00000560275.5 |
IRF9
|
interferon regulatory factor 9 |
| chr6_+_36676455 | 2.53 |
ENST00000615513.4
|
CDKN1A
|
cyclin dependent kinase inhibitor 1A |
| chr22_+_35648438 | 2.51 |
ENST00000409652.5
|
APOL6
|
apolipoprotein L6 |
| chr6_+_36676489 | 2.49 |
ENST00000448526.6
|
CDKN1A
|
cyclin dependent kinase inhibitor 1A |
| chr6_+_36678699 | 2.43 |
ENST00000405375.5
ENST00000244741.10 ENST00000373711.3 |
CDKN1A
|
cyclin dependent kinase inhibitor 1A |
| chr7_-_116030750 | 2.36 |
ENST00000265440.12
ENST00000320239.11 |
TFEC
|
transcription factor EC |
| chr2_-_201698628 | 2.34 |
ENST00000602867.1
ENST00000409474.8 |
MPP4
|
membrane palmitoylated protein 4 |
| chr19_+_56595279 | 2.33 |
ENST00000328070.10
ENST00000599599.7 ENST00000650950.1 |
ZNF71
ZIM2-AS1
|
zinc finger protein 71 ZIM2 antisense RNA 1 |
| chr2_-_201698692 | 2.27 |
ENST00000315506.11
ENST00000359962.9 ENST00000620095.4 |
MPP4
|
membrane palmitoylated protein 4 |
| chr15_+_66453418 | 2.09 |
ENST00000566326.1
|
MAP2K1
|
mitogen-activated protein kinase kinase 1 |
| chr3_-_71064915 | 2.06 |
ENST00000614176.5
ENST00000485326.7 |
FOXP1
|
forkhead box P1 |
| chr16_+_4624811 | 2.00 |
ENST00000415496.5
ENST00000262370.12 ENST00000587747.5 ENST00000399577.9 ENST00000588994.5 ENST00000586183.5 |
MGRN1
|
mahogunin ring finger 1 |
| chr3_-_71064964 | 1.82 |
ENST00000650387.1
|
FOXP1
|
forkhead box P1 |
| chr6_-_32843994 | 1.79 |
ENST00000395339.7
ENST00000374882.8 |
PSMB8
|
proteasome 20S subunit beta 8 |
| chrX_+_150693360 | 1.70 |
ENST00000370390.7
ENST00000490316.6 ENST00000542156.5 ENST00000445323.7 |
MTMR1
|
myotubularin related protein 1 |
| chr3_-_165196369 | 1.67 |
ENST00000475390.2
|
SLITRK3
|
SLIT and NTRK like family member 3 |
| chr2_+_74206384 | 1.59 |
ENST00000678623.1
ENST00000678731.1 |
MTHFD2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr12_+_20695323 | 1.47 |
ENST00000266509.7
|
SLCO1C1
|
solute carrier organic anion transporter family member 1C1 |
| chr2_-_189179754 | 1.38 |
ENST00000374866.9
ENST00000618828.1 |
COL5A2
|
collagen type V alpha 2 chain |
| chr4_+_145619371 | 1.28 |
ENST00000649156.2
ENST00000648388.1 |
MMAA
|
metabolism of cobalamin associated A |
| chr2_+_102187015 | 1.19 |
ENST00000441515.3
ENST00000264257.7 |
IL1RL2
|
interleukin 1 receptor like 2 |
| chr3_+_171843337 | 1.19 |
ENST00000334567.9
ENST00000619900.4 ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr17_+_7455571 | 1.17 |
ENST00000575379.1
|
CHRNB1
|
cholinergic receptor nicotinic beta 1 subunit |
| chr17_+_42980547 | 1.15 |
ENST00000361677.5
ENST00000589705.1 |
RUNDC1
|
RUN domain containing 1 |
| chr1_+_202462730 | 1.13 |
ENST00000290419.9
ENST00000491336.5 |
PPP1R12B
|
protein phosphatase 1 regulatory subunit 12B |
| chr3_+_38138478 | 1.12 |
ENST00000396334.8
ENST00000417037.8 ENST00000652213.1 ENST00000650112.2 ENST00000651800.2 ENST00000421516.3 ENST00000650905.2 |
MYD88
|
MYD88 innate immune signal transduction adaptor |
| chr1_-_230869564 | 1.11 |
ENST00000470540.5
|
C1orf198
|
chromosome 1 open reading frame 198 |
| chr1_+_15684284 | 1.11 |
ENST00000375799.8
ENST00000375793.2 |
PLEKHM2
|
pleckstrin homology and RUN domain containing M2 |
| chr5_+_157731400 | 1.10 |
ENST00000231198.12
|
THG1L
|
tRNA-histidine guanylyltransferase 1 like |
| chr17_-_51120855 | 1.06 |
ENST00000618113.4
ENST00000357122.8 ENST00000262013.12 |
SPAG9
|
sperm associated antigen 9 |
| chr2_+_68734773 | 1.04 |
ENST00000409202.8
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr19_+_49642188 | 1.00 |
ENST00000360565.8
|
SCAF1
|
SR-related CTD associated factor 1 |
| chr17_-_79950828 | 0.98 |
ENST00000572862.5
ENST00000573782.5 ENST00000574427.1 ENST00000570373.5 ENST00000340848.11 ENST00000576768.5 |
TBC1D16
|
TBC1 domain family member 16 |
| chr5_-_161546671 | 0.82 |
ENST00000517547.5
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr17_-_42980393 | 0.81 |
ENST00000409446.8
ENST00000409399.6 ENST00000421990.7 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 like PTGES3L-AARSD1 readthrough |
| chr5_-_161546970 | 0.80 |
ENST00000675303.1
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr4_-_139556199 | 0.80 |
ENST00000274031.8
ENST00000506866.6 |
SETD7
|
SET domain containing 7, histone lysine methyltransferase |
| chr9_-_83267230 | 0.79 |
ENST00000328788.5
|
FRMD3
|
FERM domain containing 3 |
| chr5_+_137867852 | 0.78 |
ENST00000421631.6
ENST00000239926.9 |
MYOT
|
myotilin |
| chr8_-_109334112 | 0.78 |
ENST00000678094.1
|
NUDCD1
|
NudC domain containing 1 |
| chr3_+_111542178 | 0.77 |
ENST00000283285.10
ENST00000352690.9 |
CD96
|
CD96 molecule |
| chr8_-_109334074 | 0.76 |
ENST00000239690.9
|
NUDCD1
|
NudC domain containing 1 |
| chr12_-_51009264 | 0.75 |
ENST00000545993.7
|
SLC11A2
|
solute carrier family 11 member 2 |
| chr7_+_117480011 | 0.74 |
ENST00000649406.1
ENST00000648260.1 ENST00000003084.11 |
CFTR
|
CF transmembrane conductance regulator |
| chr11_-_120138104 | 0.74 |
ENST00000341846.10
|
TRIM29
|
tripartite motif containing 29 |
| chr11_+_117200188 | 0.73 |
ENST00000529792.5
|
TAGLN
|
transgelin |
| chr4_-_186555567 | 0.73 |
ENST00000307161.5
|
MTNR1A
|
melatonin receptor 1A |
| chr22_-_31140494 | 0.72 |
ENST00000215885.4
|
PLA2G3
|
phospholipase A2 group III |
| chr18_-_27990256 | 0.72 |
ENST00000675173.1
|
CDH2
|
cadherin 2 |
| chr1_+_17580474 | 0.69 |
ENST00000375415.5
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor 10 like |
| chr1_-_28058087 | 0.66 |
ENST00000373864.5
|
EYA3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr15_-_100341899 | 0.65 |
ENST00000568565.2
ENST00000268070.9 |
ADAMTS17
|
ADAM metallopeptidase with thrombospondin type 1 motif 17 |
| chr15_+_63048535 | 0.65 |
ENST00000560959.5
|
TPM1
|
tropomyosin 1 |
| chr19_+_50025714 | 0.60 |
ENST00000598809.5
ENST00000595661.5 ENST00000391821.6 |
ZNF473
|
zinc finger protein 473 |
| chr15_+_63048436 | 0.59 |
ENST00000334895.10
ENST00000404484.9 ENST00000558910.3 ENST00000317516.12 |
TPM1
|
tropomyosin 1 |
| chr3_+_77039836 | 0.59 |
ENST00000461745.5
|
ROBO2
|
roundabout guidance receptor 2 |
| chr16_+_67227105 | 0.59 |
ENST00000563953.5
ENST00000304800.14 ENST00000565201.1 |
TMEM208
|
transmembrane protein 208 |
| chr20_+_8132138 | 0.59 |
ENST00000378641.7
ENST00000338037.11 ENST00000629992.2 |
PLCB1
|
phospholipase C beta 1 |
| chr1_+_50106265 | 0.58 |
ENST00000357083.8
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr1_-_1273617 | 0.57 |
ENST00000360466.6
|
UBE2J2
|
ubiquitin conjugating enzyme E2 J2 |
| chr21_+_34364003 | 0.55 |
ENST00000290310.4
|
KCNE2
|
potassium voltage-gated channel subfamily E regulatory subunit 2 |
| chr1_+_50105666 | 0.54 |
ENST00000651347.1
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr5_-_161546708 | 0.52 |
ENST00000393959.6
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr3_+_156120572 | 0.52 |
ENST00000389636.9
ENST00000490337.6 |
KCNAB1
|
potassium voltage-gated channel subfamily A member regulatory beta subunit 1 |
| chr12_-_6635938 | 0.52 |
ENST00000329858.9
|
LPAR5
|
lysophosphatidic acid receptor 5 |
| chr19_+_50025989 | 0.50 |
ENST00000445728.7
ENST00000270617.8 ENST00000601364.5 |
ZNF473
|
zinc finger protein 473 |
| chr3_+_111542134 | 0.50 |
ENST00000438817.6
|
CD96
|
CD96 molecule |
| chr17_+_2056073 | 0.50 |
ENST00000576444.1
ENST00000322941.3 |
HIC1
|
HIC ZBTB transcriptional repressor 1 |
| chr19_-_34677157 | 0.49 |
ENST00000601241.6
|
SCGB2B2
|
secretoglobin family 2B member 2 |
| chr3_-_108953870 | 0.48 |
ENST00000261047.8
|
GUCA1C
|
guanylate cyclase activator 1C |
| chr1_+_9239835 | 0.47 |
ENST00000602477.1
|
H6PD
|
hexose-6-phosphate dehydrogenase/glucose 1-dehydrogenase |
| chr3_+_188153271 | 0.47 |
ENST00000448637.5
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr2_+_95165784 | 0.46 |
ENST00000622059.4
ENST00000614034.5 ENST00000611147.1 |
ZNF2
|
zinc finger protein 2 |
| chr2_+_68734861 | 0.46 |
ENST00000467265.5
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr7_+_129502517 | 0.45 |
ENST00000462322.3
|
SMKR1
|
small lysine rich protein 1 |
| chr7_-_93226449 | 0.45 |
ENST00000394468.7
ENST00000453812.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr20_+_9069076 | 0.43 |
ENST00000378473.9
|
PLCB4
|
phospholipase C beta 4 |
| chr2_+_108377947 | 0.41 |
ENST00000272452.7
|
SULT1C4
|
sulfotransferase family 1C member 4 |
| chr20_+_37744630 | 0.41 |
ENST00000373473.5
|
CTNNBL1
|
catenin beta like 1 |
| chr12_+_26011713 | 0.41 |
ENST00000542004.5
|
RASSF8
|
Ras association domain family member 8 |
| chr2_+_27537380 | 0.40 |
ENST00000447166.2
|
C2orf16
|
chromosome 2 open reading frame 16 |
| chr4_-_154590735 | 0.40 |
ENST00000403106.8
ENST00000622532.1 ENST00000651975.1 |
FGA
|
fibrinogen alpha chain |
| chr3_+_45388555 | 0.39 |
ENST00000650792.2
ENST00000414984.5 ENST00000645846.2 |
LARS2
|
leucyl-tRNA synthetase 2, mitochondrial |
| chr12_-_9999176 | 0.39 |
ENST00000298527.10
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1 member B |
| chr14_+_73490926 | 0.39 |
ENST00000304061.8
|
RIOX1
|
ribosomal oxygenase 1 |
| chr4_+_157220691 | 0.39 |
ENST00000509417.5
ENST00000645636.1 ENST00000296526.12 ENST00000264426.14 |
GRIA2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr2_-_96035974 | 0.38 |
ENST00000434632.5
|
GPAT2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr15_+_63048576 | 0.38 |
ENST00000559281.6
|
TPM1
|
tropomyosin 1 |
| chr1_-_161367872 | 0.37 |
ENST00000367974.2
|
CFAP126
|
cilia and flagella associated protein 126 |
| chr15_+_63048658 | 0.37 |
ENST00000560615.5
ENST00000651577.1 |
TPM1
|
tropomyosin 1 |
| chr10_+_102644990 | 0.37 |
ENST00000645961.1
|
TRIM8
|
tripartite motif containing 8 |
| chr11_-_133845495 | 0.37 |
ENST00000299140.8
ENST00000532889.1 |
SPATA19
|
spermatogenesis associated 19 |
| chr11_-_85627268 | 0.37 |
ENST00000650630.1
|
DLG2
|
discs large MAGUK scaffold protein 2 |
| chrX_+_108045050 | 0.36 |
ENST00000458383.1
ENST00000217957.10 |
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr7_+_117480052 | 0.36 |
ENST00000426809.5
ENST00000649781.1 |
CFTR
|
CF transmembrane conductance regulator |
| chr1_+_192636121 | 0.36 |
ENST00000543215.5
ENST00000391995.7 |
RGS13
|
regulator of G protein signaling 13 |
| chr12_-_11022620 | 0.35 |
ENST00000390673.2
|
TAS2R19
|
taste 2 receptor member 19 |
| chr12_-_11092313 | 0.35 |
ENST00000531678.1
|
TAS2R43
|
taste 2 receptor member 43 |
| chr3_+_130345516 | 0.34 |
ENST00000265379.10
|
COL6A5
|
collagen type VI alpha 5 chain |
| chr4_+_157220654 | 0.34 |
ENST00000393815.6
|
GRIA2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr5_+_40909490 | 0.33 |
ENST00000313164.10
|
C7
|
complement C7 |
| chr1_-_161309961 | 0.33 |
ENST00000533357.5
ENST00000672602.2 ENST00000526189.3 |
MPZ
|
myelin protein zero |
| chr11_-_62984957 | 0.33 |
ENST00000377871.7
ENST00000360421.9 |
SLC22A6
|
solute carrier family 22 member 6 |
| chr3_-_108953762 | 0.32 |
ENST00000393963.7
ENST00000471108.1 |
GUCA1C
|
guanylate cyclase activator 1C |
| chr13_-_26221703 | 0.32 |
ENST00000381570.7
ENST00000346166.7 |
RNF6
|
ring finger protein 6 |
| chr9_-_114074969 | 0.31 |
ENST00000466610.6
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr3_-_149221811 | 0.31 |
ENST00000455472.3
ENST00000264613.11 |
CP
|
ceruloplasmin |
| chr17_+_34356472 | 0.31 |
ENST00000225844.7
|
CCL13
|
C-C motif chemokine ligand 13 |
| chr16_-_20327426 | 0.31 |
ENST00000575582.5
ENST00000341642.9 ENST00000381362.8 ENST00000572347.5 ENST00000572478.5 ENST00000302555.10 |
GP2
|
glycoprotein 2 |
| chr2_+_113127588 | 0.30 |
ENST00000409930.4
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr17_-_40054391 | 0.30 |
ENST00000394127.6
ENST00000356271.7 ENST00000394128.7 ENST00000535071.6 ENST00000580885.5 ENST00000543759.6 ENST00000537674.6 ENST00000580517.5 ENST00000578161.2 |
MED24
|
mediator complex subunit 24 |
| chr4_-_46124046 | 0.29 |
ENST00000295452.5
|
GABRG1
|
gamma-aminobutyric acid type A receptor subunit gamma1 |
| chr16_-_18926408 | 0.28 |
ENST00000446231.7
|
SMG1
|
SMG1 nonsense mediated mRNA decay associated PI3K related kinase |
| chr6_+_108656346 | 0.28 |
ENST00000540898.1
|
FOXO3
|
forkhead box O3 |
| chr6_+_31571957 | 0.28 |
ENST00000454783.5
|
LTA
|
lymphotoxin alpha |
| chr12_+_57745017 | 0.27 |
ENST00000547992.5
ENST00000552816.5 ENST00000257910.8 ENST00000547472.5 |
TSPAN31
|
tetraspanin 31 |
| chr6_-_49744378 | 0.27 |
ENST00000371159.8
ENST00000263045.9 |
CRISP3
|
cysteine rich secretory protein 3 |
| chr16_+_78022538 | 0.26 |
ENST00000651443.1
ENST00000299642.10 |
CLEC3A
|
C-type lectin domain family 3 member A |
| chr11_+_64924673 | 0.26 |
ENST00000164133.7
ENST00000532850.1 |
PPP2R5B
|
protein phosphatase 2 regulatory subunit B'beta |
| chr12_+_100357068 | 0.26 |
ENST00000323346.10
|
SLC17A8
|
solute carrier family 17 member 8 |
| chr4_-_139084289 | 0.26 |
ENST00000510408.5
ENST00000379549.7 ENST00000358635.7 |
ELF2
|
E74 like ETS transcription factor 2 |
| chr12_-_10986912 | 0.25 |
ENST00000506868.1
|
TAS2R50
|
taste 2 receptor member 50 |
| chr5_-_16508990 | 0.25 |
ENST00000399793.6
|
RETREG1
|
reticulophagy regulator 1 |
| chrX_+_108044967 | 0.25 |
ENST00000415430.7
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr20_+_31739260 | 0.25 |
ENST00000340513.4
ENST00000300403.11 |
TPX2
|
TPX2 microtubule nucleation factor |
| chr18_+_63476927 | 0.25 |
ENST00000489441.5
ENST00000382771.9 ENST00000424602.1 |
SERPINB5
|
serpin family B member 5 |
| chr12_-_91004965 | 0.25 |
ENST00000261172.8
|
EPYC
|
epiphycan |
| chrX_-_138711663 | 0.25 |
ENST00000315930.11
|
FGF13
|
fibroblast growth factor 13 |
| chr10_+_17752185 | 0.24 |
ENST00000377495.2
|
TMEM236
|
transmembrane protein 236 |
| chr13_-_40982880 | 0.23 |
ENST00000635415.1
|
ELF1
|
E74 like ETS transcription factor 1 |
| chr6_+_46793379 | 0.23 |
ENST00000230588.9
ENST00000611727.2 |
MEP1A
|
meprin A subunit alpha |
| chr6_-_25874212 | 0.23 |
ENST00000361703.10
ENST00000397060.8 |
SLC17A3
|
solute carrier family 17 member 3 |
| chr5_-_16508951 | 0.23 |
ENST00000682628.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr19_-_51372686 | 0.23 |
ENST00000595217.1
|
NKG7
|
natural killer cell granule protein 7 |
| chr3_+_15427551 | 0.23 |
ENST00000396842.7
|
EAF1
|
ELL associated factor 1 |
| chr1_-_230745574 | 0.23 |
ENST00000681269.1
|
AGT
|
angiotensinogen |
| chr5_-_16508858 | 0.22 |
ENST00000684456.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr1_+_15236509 | 0.22 |
ENST00000683790.1
|
FHAD1
|
forkhead associated phosphopeptide binding domain 1 |
| chr19_-_56393430 | 0.21 |
ENST00000589895.1
ENST00000589143.5 ENST00000301310.8 ENST00000586929.5 |
ZNF582
|
zinc finger protein 582 |
| chr3_-_196515315 | 0.21 |
ENST00000397537.3
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr1_-_33036840 | 0.21 |
ENST00000548033.5
ENST00000487289.1 ENST00000626911.1 ENST00000673291.1 ENST00000480134.5 ENST00000373449.7 ENST00000672715.1 ENST00000467905.5 ENST00000629371.2 |
AK2
|
adenylate kinase 2 |
| chr12_-_48852153 | 0.19 |
ENST00000308025.8
|
DDX23
|
DEAD-box helicase 23 |
| chr6_-_123636979 | 0.19 |
ENST00000662930.1
|
TRDN
|
triadin |
| chr16_-_46748337 | 0.18 |
ENST00000394809.9
|
MYLK3
|
myosin light chain kinase 3 |
| chr6_+_31572279 | 0.18 |
ENST00000418386.3
|
LTA
|
lymphotoxin alpha |
| chr19_-_54159696 | 0.17 |
ENST00000222224.4
|
LENG1
|
leukocyte receptor cluster member 1 |
| chr16_+_85899121 | 0.16 |
ENST00000268638.10
ENST00000565552.1 |
IRF8
|
interferon regulatory factor 8 |
| chr6_-_123636923 | 0.16 |
ENST00000334268.9
|
TRDN
|
triadin |
| chr6_-_56851888 | 0.16 |
ENST00000312431.10
ENST00000520645.5 |
DST
|
dystonin |
| chr10_+_104269163 | 0.16 |
ENST00000338595.7
|
GSTO2
|
glutathione S-transferase omega 2 |
| chrX_+_106802660 | 0.15 |
ENST00000357242.10
ENST00000310452.6 ENST00000481617.6 ENST00000276175.7 |
TBC1D8B
|
TBC1 domain family member 8B |
| chr12_+_5432101 | 0.15 |
ENST00000423158.4
|
NTF3
|
neurotrophin 3 |
| chr1_+_115976488 | 0.14 |
ENST00000369503.9
|
SLC22A15
|
solute carrier family 22 member 15 |
| chr11_+_73292755 | 0.14 |
ENST00000680955.1
ENST00000538328.2 |
P2RY6
|
pyrimidinergic receptor P2Y6 |
| chr6_+_139028680 | 0.14 |
ENST00000367660.4
|
ABRACL
|
ABRA C-terminal like |
| chr1_-_33036927 | 0.14 |
ENST00000354858.11
|
AK2
|
adenylate kinase 2 |
| chr5_+_141343818 | 0.13 |
ENST00000619750.1
ENST00000253812.8 |
PCDHGA3
|
protocadherin gamma subfamily A, 3 |
| chr11_-_120128831 | 0.13 |
ENST00000529044.5
|
TRIM29
|
tripartite motif containing 29 |
| chr11_-_118176576 | 0.13 |
ENST00000278947.6
|
SCN2B
|
sodium voltage-gated channel beta subunit 2 |
| chr14_+_96256194 | 0.13 |
ENST00000216629.11
ENST00000553356.1 |
BDKRB1
|
bradykinin receptor B1 |
| chr6_+_52186373 | 0.13 |
ENST00000648244.1
|
IL17A
|
interleukin 17A |
| chr19_-_50025936 | 0.13 |
ENST00000596445.5
ENST00000599538.5 |
VRK3
|
VRK serine/threonine kinase 3 |
| chr5_-_65481907 | 0.12 |
ENST00000381055.8
|
ADAMTS6
|
ADAM metallopeptidase with thrombospondin type 1 motif 6 |
| chr5_-_128339191 | 0.12 |
ENST00000507835.5
|
FBN2
|
fibrillin 2 |
| chr19_-_44448435 | 0.12 |
ENST00000588655.1
ENST00000592308.1 ENST00000614049.5 ENST00000613197.4 |
ENSG00000267188.1
ZNF229
|
novel transcript zinc finger protein 229 |
| chrX_-_66033664 | 0.12 |
ENST00000427538.5
|
VSIG4
|
V-set and immunoglobulin domain containing 4 |
| chr1_-_205422050 | 0.12 |
ENST00000367153.9
|
LEMD1
|
LEM domain containing 1 |
| chr5_-_16508812 | 0.12 |
ENST00000683414.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr15_+_57706670 | 0.11 |
ENST00000299638.8
|
POLR2M
|
RNA polymerase II subunit M |
| chr2_+_231056845 | 0.11 |
ENST00000677230.1
ENST00000677259.1 ENST00000677180.1 ENST00000409643.6 ENST00000619128.5 ENST00000678679.1 ENST00000676740.1 ENST00000308696.11 ENST00000440838.5 ENST00000373635.9 |
PSMD1
|
proteasome 26S subunit, non-ATPase 1 |
| chr10_+_102644887 | 0.11 |
ENST00000462202.3
|
TRIM8
|
tripartite motif containing 8 |
| chr5_+_90899183 | 0.10 |
ENST00000640815.1
|
ADGRV1
|
adhesion G protein-coupled receptor V1 |
| chr3_-_15427497 | 0.09 |
ENST00000443029.5
ENST00000383789.9 ENST00000450816.6 ENST00000383790.8 |
METTL6
|
methyltransferase like 6 |
| chr6_+_83512501 | 0.08 |
ENST00000369700.4
|
PRSS35
|
serine protease 35 |
| chr2_-_201769759 | 0.08 |
ENST00000680163.1
ENST00000680188.1 ENST00000679550.1 |
ALS2
|
alsin Rho guanine nucleotide exchange factor ALS2 |
| chr13_+_111115303 | 0.08 |
ENST00000646102.2
ENST00000449979.5 |
ARHGEF7
|
Rho guanine nucleotide exchange factor 7 |
| chr6_+_25754699 | 0.08 |
ENST00000439485.6
ENST00000377905.9 |
SLC17A4
|
solute carrier family 17 member 4 |
| chr19_+_44113385 | 0.07 |
ENST00000262894.11
ENST00000588926.5 ENST00000592780.5 |
ZNF225
|
zinc finger protein 225 |
| chr6_+_133889105 | 0.06 |
ENST00000367882.5
|
TCF21
|
transcription factor 21 |
| chr5_-_131945658 | 0.05 |
ENST00000442687.6
ENST00000616644.2 |
MEIKIN
|
meiotic kinetochore factor |
| chr10_+_113553039 | 0.04 |
ENST00000351270.4
|
HABP2
|
hyaluronan binding protein 2 |
| chr3_+_186581995 | 0.03 |
ENST00000418776.1
|
ENSG00000283149.1
|
novel protein |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 13.3 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 1.6 | 6.3 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 1.5 | 7.5 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 1.2 | 5.0 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 1.1 | 6.8 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.9 | 10.0 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.9 | 5.3 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.8 | 7.5 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.7 | 4.3 | GO:0021564 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) vagus nerve development(GO:0021564) |
| 0.5 | 1.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.4 | 1.3 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.4 | 1.7 | GO:1902159 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.4 | 2.0 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.4 | 4.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.3 | 1.3 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.3 | 2.1 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.2 | 4.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.2 | 3.9 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.2 | 1.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.2 | 2.0 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.2 | 3.1 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.2 | 0.5 | GO:0002876 | positive regulation of chronic inflammatory response to antigenic stimulus(GO:0002876) |
| 0.2 | 1.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.2 | 0.8 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.1 | 0.6 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.1 | 1.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.3 | GO:1905237 | response to cyclosporin A(GO:1905237) |
| 0.1 | 0.8 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 0.4 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.1 | 1.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.3 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 1.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.3 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.1 | 0.2 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 0.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.7 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 2.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 8.5 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.1 | 0.2 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.1 | 0.3 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.1 | 1.5 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.1 | 0.2 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.1 | 0.3 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.7 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 1.5 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.8 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 1.6 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.0 | 0.7 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.4 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 1.7 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.3 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.5 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.1 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.6 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.3 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.5 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.4 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 4.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.2 | GO:0009407 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 0.0 | 0.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.5 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 1.0 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 2.8 | GO:0071357 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.5 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.4 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.1 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.5 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.0 | 1.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.6 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 1.3 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.4 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 2.8 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 13.3 | GO:0042825 | TAP complex(GO:0042825) |
| 1.5 | 7.5 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 1.2 | 9.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.8 | 6.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.5 | 1.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 0.7 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.2 | 1.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.2 | 4.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 0.8 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 2.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.3 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 2.1 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 3.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 1.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 4.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.8 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.3 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.7 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 1.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 1.1 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 10.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.7 | 13.3 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) MHC class Ib protein binding(GO:0023029) TAP2 binding(GO:0046979) |
| 1.5 | 7.5 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 1.1 | 6.8 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 1.0 | 5.0 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) CCR1 chemokine receptor binding(GO:0031726) |
| 0.7 | 4.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.6 | 6.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.5 | 4.2 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.4 | 1.6 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.4 | 1.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.3 | 8.7 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.3 | 1.2 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.3 | 1.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.2 | 0.7 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.2 | 2.1 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.2 | 0.5 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.2 | 1.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 5.3 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.2 | 0.8 | GO:0015639 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.8 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 0.3 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 1.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.3 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 1.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 3.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.7 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.4 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 2.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.3 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.3 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 2.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.7 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 4.3 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.6 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 1.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.4 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.3 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.4 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.6 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 6.2 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 0.9 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 1.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.2 | GO:0045174 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 1.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 1.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 2.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.5 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 4.1 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 1.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 5.2 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 2.6 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 6.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 6.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 9.6 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 6.4 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 6.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 13.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 4.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 5.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 4.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 3.4 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.6 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.8 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.1 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.3 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.8 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 13.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.3 | 7.5 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.3 | 4.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.2 | 5.8 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 14.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 7.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 6.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 8.5 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.1 | 1.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.1 | 1.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 1.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 1.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 2.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 2.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.7 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |