Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
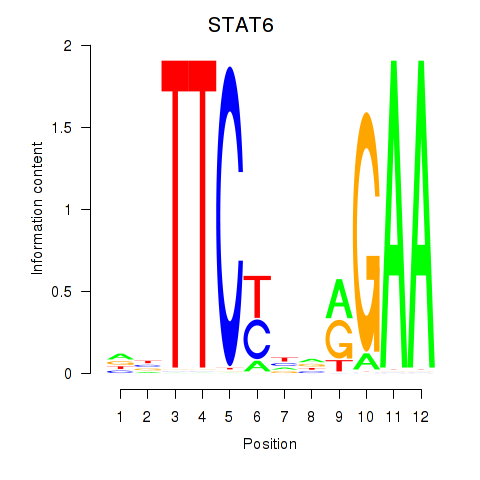
Results for STAT6
Z-value: 0.55
Transcription factors associated with STAT6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STAT6
|
ENSG00000166888.12 | STAT6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| STAT6 | hg38_v1_chr12_-_57110284_57110340, hg38_v1_chr12_-_57129001_57129100 | 0.59 | 2.0e-03 | Click! |
Activity profile of STAT6 motif
Sorted Z-values of STAT6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of STAT6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_40862354 | 1.91 |
ENST00000372638.4
|
CITED4
|
Cbp/p300 interacting transactivator with Glu/Asp rich carboxy-terminal domain 4 |
| chr11_-_128587551 | 1.62 |
ENST00000392668.8
|
ETS1
|
ETS proto-oncogene 1, transcription factor |
| chr5_+_119356011 | 1.38 |
ENST00000504771.3
ENST00000415806.2 |
TNFAIP8
|
TNF alpha induced protein 8 |
| chr1_-_120100688 | 1.12 |
ENST00000652264.1
|
NOTCH2
|
notch receptor 2 |
| chr6_-_32853618 | 0.93 |
ENST00000354258.5
|
TAP1
|
transporter 1, ATP binding cassette subfamily B member |
| chr6_-_32853813 | 0.92 |
ENST00000643049.2
|
TAP1
|
transporter 1, ATP binding cassette subfamily B member |
| chr4_-_115113822 | 0.78 |
ENST00000613194.4
|
NDST4
|
N-deacetylase and N-sulfotransferase 4 |
| chr6_+_137867241 | 0.78 |
ENST00000612899.5
ENST00000420009.5 |
TNFAIP3
|
TNF alpha induced protein 3 |
| chr6_+_32854179 | 0.69 |
ENST00000374859.3
|
PSMB9
|
proteasome 20S subunit beta 9 |
| chr1_+_159437845 | 0.67 |
ENST00000642080.1
|
OR10J1
|
olfactory receptor family 10 subfamily J member 1 |
| chr11_-_68213577 | 0.61 |
ENST00000402789.5
ENST00000402185.6 ENST00000458496.1 |
KMT5B
|
lysine methyltransferase 5B |
| chr14_-_24609660 | 0.60 |
ENST00000557220.6
ENST00000216338.9 ENST00000382548.4 |
GZMH
|
granzyme H |
| chr1_-_205422050 | 0.57 |
ENST00000367153.9
|
LEMD1
|
LEM domain containing 1 |
| chr5_+_35856883 | 0.55 |
ENST00000506850.5
ENST00000303115.8 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr17_+_34255274 | 0.54 |
ENST00000580907.5
ENST00000225831.4 |
CCL2
|
C-C motif chemokine ligand 2 |
| chr8_+_103298836 | 0.54 |
ENST00000523739.5
ENST00000358755.5 |
FZD6
|
frizzled class receptor 6 |
| chr15_+_84981981 | 0.50 |
ENST00000339708.9
|
PDE8A
|
phosphodiesterase 8A |
| chr8_+_122781621 | 0.50 |
ENST00000314393.6
|
ZHX2
|
zinc fingers and homeoboxes 2 |
| chr2_-_203535253 | 0.49 |
ENST00000457812.5
ENST00000319170.10 ENST00000630330.2 ENST00000308091.8 ENST00000453034.5 ENST00000420371.2 |
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr2_-_119366757 | 0.47 |
ENST00000414534.1
|
C2orf76
|
chromosome 2 open reading frame 76 |
| chr12_-_6342066 | 0.47 |
ENST00000162749.7
ENST00000440083.6 |
TNFRSF1A
|
TNF receptor superfamily member 1A |
| chr15_+_66453418 | 0.47 |
ENST00000566326.1
|
MAP2K1
|
mitogen-activated protein kinase kinase 1 |
| chr6_+_26440472 | 0.46 |
ENST00000494393.5
ENST00000482451.5 ENST00000471353.5 ENST00000361232.7 ENST00000487627.5 ENST00000496719.1 ENST00000244519.7 ENST00000490254.5 ENST00000487272.1 |
BTN3A3
|
butyrophilin subfamily 3 member A3 |
| chr5_-_141958174 | 0.43 |
ENST00000231484.4
|
PCDH12
|
protocadherin 12 |
| chr2_-_119366682 | 0.43 |
ENST00000409877.5
ENST00000409523.1 ENST00000409466.6 |
C2orf76
|
chromosome 2 open reading frame 76 |
| chr15_+_84981834 | 0.42 |
ENST00000394553.6
|
PDE8A
|
phosphodiesterase 8A |
| chr3_+_41194741 | 0.41 |
ENST00000643541.1
ENST00000426215.5 ENST00000645210.1 ENST00000646381.1 ENST00000405570.6 ENST00000642248.1 ENST00000433400.6 |
CTNNB1
|
catenin beta 1 |
| chr2_-_119366807 | 0.41 |
ENST00000334816.12
|
C2orf76
|
chromosome 2 open reading frame 76 |
| chr12_-_51009264 | 0.40 |
ENST00000545993.7
|
SLC11A2
|
solute carrier family 11 member 2 |
| chr5_-_147081428 | 0.39 |
ENST00000394413.7
|
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
| chr4_+_95051671 | 0.37 |
ENST00000440890.7
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr6_-_11232658 | 0.37 |
ENST00000379433.5
ENST00000379446.10 ENST00000620854.4 |
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr20_-_18497218 | 0.37 |
ENST00000337227.9
|
RBBP9
|
RB binding protein 9, serine hydrolase |
| chr15_-_100341899 | 0.35 |
ENST00000568565.2
ENST00000268070.9 |
ADAMTS17
|
ADAM metallopeptidase with thrombospondin type 1 motif 17 |
| chr17_-_41350824 | 0.33 |
ENST00000007735.4
|
KRT33A
|
keratin 33A |
| chr14_+_22052503 | 0.33 |
ENST00000390449.3
|
TRAV21
|
T cell receptor alpha variable 21 |
| chr3_+_141402322 | 0.31 |
ENST00000510338.5
ENST00000504673.5 |
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr18_+_58196736 | 0.31 |
ENST00000675221.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr4_+_54100161 | 0.31 |
ENST00000326902.7
ENST00000503800.1 |
GSX2
|
GS homeobox 2 |
| chr1_+_26432803 | 0.30 |
ENST00000430232.5
|
DHDDS
|
dehydrodolichyl diphosphate synthase subunit |
| chr11_+_73647549 | 0.28 |
ENST00000227214.10
ENST00000398494.8 ENST00000543085.5 |
PLEKHB1
|
pleckstrin homology domain containing B1 |
| chr17_-_39778213 | 0.28 |
ENST00000583368.1
|
IKZF3
|
IKAROS family zinc finger 3 |
| chr10_+_100347225 | 0.28 |
ENST00000370355.3
|
SCD
|
stearoyl-CoA desaturase |
| chr4_+_72031902 | 0.27 |
ENST00000344413.6
ENST00000308744.12 |
NPFFR2
|
neuropeptide FF receptor 2 |
| chrX_-_135764444 | 0.27 |
ENST00000597510.6
|
CT45A3
|
cancer/testis antigen family 45 member A3 |
| chr16_-_74421756 | 0.26 |
ENST00000617101.4
|
CLEC18B
|
C-type lectin domain family 18 member B |
| chr6_-_24877262 | 0.26 |
ENST00000378023.8
ENST00000540914.5 |
RIPOR2
|
RHO family interacting cell polarization regulator 2 |
| chr11_+_112961402 | 0.25 |
ENST00000613217.4
ENST00000316851.12 ENST00000620046.4 ENST00000531044.5 ENST00000529356.5 |
NCAM1
|
neural cell adhesion molecule 1 |
| chr14_+_21918161 | 0.24 |
ENST00000390439.2
|
TRAV13-2
|
T cell receptor alpha variable 13-2 |
| chr2_+_27537380 | 0.24 |
ENST00000447166.2
|
C2orf16
|
chromosome 2 open reading frame 16 |
| chr16_-_67660694 | 0.24 |
ENST00000219251.13
ENST00000620338.4 |
ACD
|
ACD shelterin complex subunit and telomerase recruitment factor |
| chr2_+_68734861 | 0.23 |
ENST00000467265.5
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr13_-_51845169 | 0.23 |
ENST00000627246.3
ENST00000629372.3 |
TMEM272
|
transmembrane protein 272 |
| chr11_-_60952134 | 0.23 |
ENST00000679573.1
ENST00000681882.1 ENST00000681951.1 ENST00000227880.8 |
SLC15A3
|
solute carrier family 15 member 3 |
| chr17_+_7558296 | 0.23 |
ENST00000438470.5
ENST00000436057.5 |
TNFSF13
|
TNF superfamily member 13 |
| chr5_-_147081462 | 0.23 |
ENST00000508267.5
ENST00000504198.5 |
PPP2R2B
|
protein phosphatase 2 regulatory subunit Bbeta |
| chr12_+_12611839 | 0.22 |
ENST00000228865.3
|
CREBL2
|
cAMP responsive element binding protein like 2 |
| chr16_-_74421392 | 0.22 |
ENST00000339953.9
ENST00000620745.1 ENST00000682950.1 |
CLEC18B
|
C-type lectin domain family 18 member B |
| chr4_-_46390039 | 0.22 |
ENST00000540012.5
|
GABRA2
|
gamma-aminobutyric acid type A receptor subunit alpha2 |
| chr2_+_119366924 | 0.21 |
ENST00000535757.5
ENST00000535617.5 ENST00000627093.2 ENST00000355857.8 ENST00000409094.5 ENST00000542275.5 ENST00000311521.8 |
DBI
|
diazepam binding inhibitor, acyl-CoA binding protein |
| chr17_+_36103819 | 0.21 |
ENST00000615863.2
ENST00000621626.1 |
CCL4
|
C-C motif chemokine ligand 4 |
| chr11_-_60952559 | 0.21 |
ENST00000538739.2
|
SLC15A3
|
solute carrier family 15 member 3 |
| chr19_-_42442938 | 0.21 |
ENST00000601181.6
|
CXCL17
|
C-X-C motif chemokine ligand 17 |
| chr6_-_27912396 | 0.21 |
ENST00000303324.4
|
OR2B2
|
olfactory receptor family 2 subfamily B member 2 |
| chr19_-_54100792 | 0.20 |
ENST00000391761.5
ENST00000356532.7 ENST00000616447.4 ENST00000359649.8 ENST00000358375.8 ENST00000391760.1 ENST00000351806.8 |
OSCAR
|
osteoclast associated Ig-like receptor |
| chr7_+_139133744 | 0.20 |
ENST00000430935.5
ENST00000495038.5 ENST00000474035.6 ENST00000478836.6 ENST00000464848.5 ENST00000343187.8 |
TTC26
|
tetratricopeptide repeat domain 26 |
| chr2_-_174597728 | 0.20 |
ENST00000409891.5
ENST00000410117.5 |
WIPF1
|
WAS/WASL interacting protein family member 1 |
| chr2_-_202870761 | 0.20 |
ENST00000420558.5
ENST00000418208.5 |
ICA1L
|
islet cell autoantigen 1 like |
| chr16_+_69950705 | 0.20 |
ENST00000615430.4
|
CLEC18A
|
C-type lectin domain family 18 member A |
| chr8_+_96493803 | 0.20 |
ENST00000518385.5
ENST00000302190.9 |
SDC2
|
syndecan 2 |
| chr11_+_112961480 | 0.19 |
ENST00000621850.4
|
NCAM1
|
neural cell adhesion molecule 1 |
| chr16_+_69950907 | 0.19 |
ENST00000393701.6
ENST00000568461.5 |
CLEC18A
|
C-type lectin domain family 18 member A |
| chr17_-_8867639 | 0.18 |
ENST00000619866.5
|
PIK3R6
|
phosphoinositide-3-kinase regulatory subunit 6 |
| chr12_-_81758641 | 0.18 |
ENST00000552948.5
ENST00000548586.5 |
PPFIA2
|
PTPRF interacting protein alpha 2 |
| chr2_+_68734773 | 0.18 |
ENST00000409202.8
|
ARHGAP25
|
Rho GTPase activating protein 25 |
| chr11_+_65314853 | 0.18 |
ENST00000279249.3
|
CDC42EP2
|
CDC42 effector protein 2 |
| chr8_+_39934955 | 0.18 |
ENST00000502986.2
|
IDO2
|
indoleamine 2,3-dioxygenase 2 |
| chr6_-_27893175 | 0.17 |
ENST00000359611.4
|
H2AC17
|
H2A clustered histone 17 |
| chr6_-_41286665 | 0.17 |
ENST00000589614.5
ENST00000244709.9 ENST00000334475.10 ENST00000591620.1 |
TREM1
|
triggering receptor expressed on myeloid cells 1 |
| chr7_+_23598144 | 0.17 |
ENST00000410069.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr9_+_36572854 | 0.17 |
ENST00000543751.5
ENST00000536860.5 ENST00000541717.4 ENST00000536329.5 ENST00000536987.5 ENST00000298048.7 ENST00000495529.5 ENST00000545008.5 |
MELK
|
maternal embryonic leucine zipper kinase |
| chr1_-_153312919 | 0.17 |
ENST00000683862.1
|
PGLYRP3
|
peptidoglycan recognition protein 3 |
| chr1_-_100178215 | 0.16 |
ENST00000370138.1
ENST00000370137.6 ENST00000342895.7 ENST00000620882.4 |
LRRC39
|
leucine rich repeat containing 39 |
| chr4_-_120066777 | 0.16 |
ENST00000296509.11
|
MAD2L1
|
mitotic arrest deficient 2 like 1 |
| chr7_-_75772172 | 0.16 |
ENST00000005180.9
|
CCL26
|
C-C motif chemokine ligand 26 |
| chr17_+_38925168 | 0.16 |
ENST00000583195.2
|
LINC00672
|
long intergenic non-protein coding RNA 672 |
| chr1_+_42153399 | 0.16 |
ENST00000372581.2
|
GUCA2B
|
guanylate cyclase activator 2B |
| chr11_-_60952067 | 0.16 |
ENST00000681275.1
|
SLC15A3
|
solute carrier family 15 member 3 |
| chr4_-_10684749 | 0.15 |
ENST00000226951.11
|
CLNK
|
cytokine dependent hematopoietic cell linker |
| chr16_+_69951180 | 0.15 |
ENST00000288040.10
ENST00000449317.6 |
CLEC18A
|
C-type lectin domain family 18 member A |
| chrX_-_103064164 | 0.15 |
ENST00000372728.4
|
BEX1
|
brain expressed X-linked 1 |
| chr9_-_132079856 | 0.15 |
ENST00000651555.1
ENST00000651950.1 ENST00000357028.6 ENST00000474263.1 ENST00000292035.10 |
MED27
|
mediator complex subunit 27 |
| chr17_+_7558712 | 0.15 |
ENST00000338784.9
ENST00000625791.2 |
TNFSF13
|
TNF superfamily member 13 |
| chr5_-_84384871 | 0.15 |
ENST00000296591.10
|
EDIL3
|
EGF like repeats and discoidin domains 3 |
| chr11_+_112961247 | 0.14 |
ENST00000621518.4
ENST00000618266.4 ENST00000615112.4 ENST00000615285.4 ENST00000619839.4 ENST00000401611.6 ENST00000621128.4 |
NCAM1
|
neural cell adhesion molecule 1 |
| chr1_+_213989691 | 0.14 |
ENST00000607425.1
|
PROX1
|
prospero homeobox 1 |
| chr17_+_7558465 | 0.14 |
ENST00000349228.8
|
TNFSF13
|
TNF superfamily member 13 |
| chr16_+_70173783 | 0.14 |
ENST00000541793.7
ENST00000314151.12 ENST00000565806.5 ENST00000569347.6 ENST00000536907.2 |
CLEC18C
|
C-type lectin domain family 18 member C |
| chr15_-_29822077 | 0.14 |
ENST00000677774.1
|
TJP1
|
tight junction protein 1 |
| chr14_+_67720842 | 0.14 |
ENST00000267502.3
|
RDH12
|
retinol dehydrogenase 12 |
| chr17_+_7558774 | 0.14 |
ENST00000396545.4
|
TNFSF13
|
TNF superfamily member 13 |
| chr2_-_152175715 | 0.14 |
ENST00000263904.5
|
STAM2
|
signal transducing adaptor molecule 2 |
| chr6_+_10555787 | 0.13 |
ENST00000316170.9
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2 (I blood group) |
| chr12_-_81758665 | 0.13 |
ENST00000549325.5
ENST00000550584.6 |
PPFIA2
|
PTPRF interacting protein alpha 2 |
| chr8_+_10095704 | 0.13 |
ENST00000382490.9
|
MSRA
|
methionine sulfoxide reductase A |
| chr8_+_10095551 | 0.13 |
ENST00000522907.5
ENST00000528246.5 |
MSRA
|
methionine sulfoxide reductase A |
| chr12_-_9999176 | 0.13 |
ENST00000298527.10
ENST00000348658.4 |
CLEC1B
|
C-type lectin domain family 1 member B |
| chr7_+_129375643 | 0.12 |
ENST00000490911.5
|
AHCYL2
|
adenosylhomocysteinase like 2 |
| chr17_-_17836973 | 0.12 |
ENST00000261646.11
ENST00000355815.8 |
SREBF1
|
sterol regulatory element binding transcription factor 1 |
| chr1_-_203229660 | 0.12 |
ENST00000255427.7
ENST00000367229.6 |
CHIT1
|
chitinase 1 |
| chr1_+_149782671 | 0.11 |
ENST00000444948.5
ENST00000369168.5 |
FCGR1A
|
Fc fragment of IgG receptor Ia |
| chr5_-_74866958 | 0.11 |
ENST00000389156.9
|
FAM169A
|
family with sequence similarity 169 member A |
| chr13_+_48653921 | 0.11 |
ENST00000682523.1
|
CYSLTR2
|
cysteinyl leukotriene receptor 2 |
| chr19_-_7702139 | 0.11 |
ENST00000346664.9
|
FCER2
|
Fc fragment of IgE receptor II |
| chr19_-_7702124 | 0.11 |
ENST00000597921.6
|
FCER2
|
Fc fragment of IgE receptor II |
| chr13_+_50015438 | 0.11 |
ENST00000312942.2
|
KCNRG
|
potassium channel regulator |
| chr13_+_50015254 | 0.11 |
ENST00000360473.8
|
KCNRG
|
potassium channel regulator |
| chr3_-_185938006 | 0.11 |
ENST00000342294.4
ENST00000453386.7 ENST00000382191.4 |
TRA2B
|
transformer 2 beta homolog |
| chr7_-_77199808 | 0.11 |
ENST00000248598.6
|
FGL2
|
fibrinogen like 2 |
| chr15_-_29822028 | 0.10 |
ENST00000545208.6
|
TJP1
|
tight junction protein 1 |
| chr13_+_73054969 | 0.10 |
ENST00000539231.5
|
KLF5
|
Kruppel like factor 5 |
| chr2_-_201451446 | 0.10 |
ENST00000332624.8
ENST00000430254.1 |
TRAK2
|
trafficking kinesin protein 2 |
| chr6_+_83512501 | 0.10 |
ENST00000369700.4
|
PRSS35
|
serine protease 35 |
| chr10_-_59362584 | 0.10 |
ENST00000618427.4
ENST00000611933.4 |
FAM13C
|
family with sequence similarity 13 member C |
| chr10_-_59362460 | 0.10 |
ENST00000422313.6
ENST00000435852.6 ENST00000614220.4 ENST00000618804.5 ENST00000621119.4 |
FAM13C
|
family with sequence similarity 13 member C |
| chr1_-_241519701 | 0.10 |
ENST00000366560.4
ENST00000683521.1 |
FH
|
fumarate hydratase |
| chr17_+_7558258 | 0.09 |
ENST00000483039.5
ENST00000380535.8 ENST00000396542.5 |
TNFSF13
|
TNF superfamily member 13 |
| chr9_-_133121228 | 0.09 |
ENST00000372050.8
|
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr1_-_153348825 | 0.09 |
ENST00000368739.3
ENST00000359650.10 |
PGLYRP4
|
peptidoglycan recognition protein 4 |
| chr3_-_57199938 | 0.09 |
ENST00000473921.2
ENST00000295934.8 |
HESX1
|
HESX homeobox 1 |
| chr16_-_67719300 | 0.09 |
ENST00000602279.2
ENST00000268797.12 ENST00000602855.2 ENST00000602377.1 |
GFOD2
|
glucose-fructose oxidoreductase domain containing 2 |
| chr1_+_174447944 | 0.09 |
ENST00000367685.5
|
GPR52
|
G protein-coupled receptor 52 |
| chr2_-_133568393 | 0.09 |
ENST00000317721.10
ENST00000405974.7 ENST00000409261.6 ENST00000409213.5 |
NCKAP5
|
NCK associated protein 5 |
| chr2_-_26478032 | 0.09 |
ENST00000338581.10
ENST00000402415.8 |
OTOF
|
otoferlin |
| chr8_-_116874746 | 0.09 |
ENST00000297338.7
|
RAD21
|
RAD21 cohesin complex component |
| chr17_-_36534883 | 0.09 |
ENST00000620640.4
|
MYO19
|
myosin XIX |
| chr17_+_29941605 | 0.09 |
ENST00000394835.7
|
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr12_-_81759307 | 0.09 |
ENST00000547623.5
ENST00000549396.6 |
PPFIA2
|
PTPRF interacting protein alpha 2 |
| chr17_+_36211055 | 0.08 |
ENST00000617405.5
ENST00000617416.4 ENST00000613173.4 ENST00000620732.4 ENST00000620098.4 ENST00000620576.4 ENST00000620055.4 ENST00000610565.4 ENST00000620250.1 |
CCL4L2
|
C-C motif chemokine ligand 4 like 2 |
| chr4_+_183099244 | 0.08 |
ENST00000403733.8
|
WWC2
|
WW and C2 domain containing 2 |
| chr1_-_160646958 | 0.08 |
ENST00000538290.2
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1 |
| chr17_-_36534841 | 0.08 |
ENST00000614623.5
ENST00000621344.4 |
MYO19
|
myosin XIX |
| chr6_+_30557287 | 0.08 |
ENST00000376560.8
|
PRR3
|
proline rich 3 |
| chr2_-_26478113 | 0.08 |
ENST00000339598.8
|
OTOF
|
otoferlin |
| chrX_+_136169833 | 0.08 |
ENST00000628032.2
|
FHL1
|
four and a half LIM domains 1 |
| chr7_+_55365317 | 0.08 |
ENST00000254770.3
|
LANCL2
|
LanC like 2 |
| chr4_+_165378998 | 0.08 |
ENST00000402744.9
|
CPE
|
carboxypeptidase E |
| chr11_-_59212869 | 0.07 |
ENST00000361050.4
|
MPEG1
|
macrophage expressed 1 |
| chr2_-_202871433 | 0.07 |
ENST00000457524.5
ENST00000421334.1 ENST00000617388.4 |
ICA1L
|
islet cell autoantigen 1 like |
| chr2_-_189179754 | 0.07 |
ENST00000374866.9
ENST00000618828.1 |
COL5A2
|
collagen type V alpha 2 chain |
| chrX_-_139832235 | 0.07 |
ENST00000327569.7
ENST00000361648.6 |
ATP11C
|
ATPase phospholipid transporting 11C |
| chr5_+_150671588 | 0.07 |
ENST00000523553.1
|
MYOZ3
|
myozenin 3 |
| chr4_+_6693870 | 0.07 |
ENST00000296370.4
|
S100P
|
S100 calcium binding protein P |
| chr22_-_29061831 | 0.07 |
ENST00000216071.5
|
C22orf31
|
chromosome 22 open reading frame 31 |
| chr5_+_127649018 | 0.07 |
ENST00000379445.7
|
CTXN3
|
cortexin 3 |
| chr17_+_36210924 | 0.07 |
ENST00000615418.4
|
CCL4L2
|
C-C motif chemokine ligand 4 like 2 |
| chr15_+_78873723 | 0.07 |
ENST00000559690.5
ENST00000559158.5 |
MORF4L1
|
mortality factor 4 like 1 |
| chr12_-_32755876 | 0.07 |
ENST00000324868.13
|
YARS2
|
tyrosyl-tRNA synthetase 2 |
| chr18_+_13277351 | 0.06 |
ENST00000679091.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr15_+_91100194 | 0.06 |
ENST00000394232.6
|
SV2B
|
synaptic vesicle glycoprotein 2B |
| chr12_+_51888217 | 0.06 |
ENST00000340970.8
|
ANKRD33
|
ankyrin repeat domain 33 |
| chr12_+_8992029 | 0.06 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin like receptor G1 |
| chr1_+_202462730 | 0.06 |
ENST00000290419.9
ENST00000491336.5 |
PPP1R12B
|
protein phosphatase 1 regulatory subunit 12B |
| chr12_+_51888083 | 0.06 |
ENST00000301190.11
|
ANKRD33
|
ankyrin repeat domain 33 |
| chr6_+_30557274 | 0.06 |
ENST00000376557.3
|
PRR3
|
proline rich 3 |
| chr7_-_139716980 | 0.06 |
ENST00000342645.7
|
HIPK2
|
homeodomain interacting protein kinase 2 |
| chr8_-_81447428 | 0.06 |
ENST00000256103.3
ENST00000519260.1 |
PMP2
|
peripheral myelin protein 2 |
| chr8_-_30912998 | 0.06 |
ENST00000643185.2
|
TEX15
|
testis expressed 15, meiosis and synapsis associated |
| chr1_-_8026283 | 0.06 |
ENST00000474874.5
ENST00000469499.5 ENST00000377482.10 |
ERRFI1
|
ERBB receptor feedback inhibitor 1 |
| chr1_-_201946469 | 0.06 |
ENST00000367288.5
|
LMOD1
|
leiomodin 1 |
| chr1_+_34166883 | 0.05 |
ENST00000373374.7
|
C1orf94
|
chromosome 1 open reading frame 94 |
| chrX_+_136169891 | 0.05 |
ENST00000449474.5
|
FHL1
|
four and a half LIM domains 1 |
| chr17_+_7481332 | 0.05 |
ENST00000412468.3
|
SLC35G6
|
solute carrier family 35 member G6 |
| chr5_+_139273752 | 0.05 |
ENST00000509990.5
ENST00000506147.5 ENST00000512107.5 |
MATR3
|
matrin 3 |
| chr18_-_26865689 | 0.05 |
ENST00000675739.1
ENST00000383168.9 ENST00000672981.2 ENST00000578776.1 |
AQP4
|
aquaporin 4 |
| chr17_-_3298360 | 0.05 |
ENST00000323404.2
|
OR3A1
|
olfactory receptor family 3 subfamily A member 1 |
| chr4_-_46993520 | 0.05 |
ENST00000264318.4
|
GABRA4
|
gamma-aminobutyric acid type A receptor subunit alpha4 |
| chrX_+_136169664 | 0.05 |
ENST00000456445.5
|
FHL1
|
four and a half LIM domains 1 |
| chr2_-_60550900 | 0.05 |
ENST00000643222.1
ENST00000643459.1 ENST00000489516.7 |
BCL11A
|
BAF chromatin remodeling complex subunit BCL11A |
| chr11_+_70078291 | 0.05 |
ENST00000355303.9
|
ANO1
|
anoctamin 1 |
| chr14_-_37172553 | 0.05 |
ENST00000331299.6
|
SLC25A21
|
solute carrier family 25 member 21 |
| chr2_-_202870862 | 0.05 |
ENST00000454326.5
ENST00000432273.5 ENST00000450143.5 ENST00000411681.5 |
ICA1L
|
islet cell autoantigen 1 like |
| chr9_-_70869076 | 0.04 |
ENST00000677594.1
|
TRPM3
|
transient receptor potential cation channel subfamily M member 3 |
| chr15_+_64094060 | 0.04 |
ENST00000560829.5
|
SNX1
|
sorting nexin 1 |
| chr20_+_59676661 | 0.04 |
ENST00000355648.8
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr4_-_46390273 | 0.04 |
ENST00000515082.5
|
GABRA2
|
gamma-aminobutyric acid type A receptor subunit alpha2 |
| chr1_-_160647287 | 0.04 |
ENST00000235739.6
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1 |
| chr16_+_30064462 | 0.03 |
ENST00000412304.6
|
ALDOA
|
aldolase, fructose-bisphosphate A |
| chr3_+_35641421 | 0.03 |
ENST00000449196.5
|
ARPP21
|
cAMP regulated phosphoprotein 21 |
| chr6_+_31571957 | 0.03 |
ENST00000454783.5
|
LTA
|
lymphotoxin alpha |
| chr15_+_34345865 | 0.03 |
ENST00000333756.4
|
NUTM1
|
NUT midline carcinoma family member 1 |
| chr3_-_98517096 | 0.03 |
ENST00000513873.1
|
CLDND1
|
claudin domain containing 1 |
| chr11_+_8019193 | 0.03 |
ENST00000534099.5
|
TUB
|
TUB bipartite transcription factor |
| chr4_-_46390100 | 0.03 |
ENST00000381620.9
|
GABRA2
|
gamma-aminobutyric acid type A receptor subunit alpha2 |
| chr2_+_113127588 | 0.03 |
ENST00000409930.4
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr2_-_151971750 | 0.02 |
ENST00000636598.1
|
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr18_-_26865732 | 0.02 |
ENST00000672188.1
|
AQP4
|
aquaporin 4 |
| chr15_+_24954912 | 0.02 |
ENST00000584968.5
ENST00000346403.10 ENST00000554227.6 ENST00000390687.9 ENST00000579070.5 ENST00000577565.1 ENST00000577949.5 ENST00000338327.4 |
SNRPN
SNURF
|
small nuclear ribonucleoprotein polypeptide N SNRPN upstream reading frame |
| chr10_-_95693893 | 0.02 |
ENST00000371209.5
ENST00000680144.1 ENST00000430368.6 ENST00000371217.10 |
TCTN3
|
tectonic family member 3 |
| chr5_+_7396099 | 0.02 |
ENST00000338316.9
|
ADCY2
|
adenylate cyclase 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.3 | 0.8 | GO:0034148 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.2 | 0.5 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.1 | 1.6 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.1 | 0.5 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.1 | 0.3 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.1 | 0.3 | GO:0051714 | positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 | 0.4 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.1 | 0.4 | GO:1904793 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.1 | 0.5 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.1 | 1.1 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.5 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 0.6 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 0.8 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.2 | GO:1904744 | positive regulation of telomeric DNA binding(GO:1904744) |
| 0.1 | 0.4 | GO:1902731 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.1 | 0.3 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.1 | 0.3 | GO:1903904 | negative regulation of establishment of T cell polarity(GO:1903904) negative regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001107) |
| 0.0 | 0.7 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.1 | GO:2000979 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.0 | 0.2 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.0 | 0.2 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 0.3 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.2 | GO:2000501 | regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.0 | 0.5 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.1 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.0 | 0.9 | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.6 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.2 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.2 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) |
| 0.0 | 0.0 | GO:1904800 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.1 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.6 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:0030578 | PML body organization(GO:0030578) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.7 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.4 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.4 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.2 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:0060342 | photoreceptor inner segment membrane(GO:0060342) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.3 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 0.5 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) MHC class Ib protein binding(GO:0023029) TAP2 binding(GO:0046979) |
| 0.1 | 0.5 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 1.1 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.1 | 0.8 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.3 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.1 | 0.6 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.1 | 0.4 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.5 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.1 | 0.2 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.1 | 0.3 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 0.3 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.1 | 0.2 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.1 | 0.3 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.5 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.2 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.2 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 1.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.7 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 2.0 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 1.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.7 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.0 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.2 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 1.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.5 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.8 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.5 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.4 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |