Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for TBX19
Z-value: 0.44
Transcription factors associated with TBX19
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX19
|
ENSG00000143178.13 | TBX19 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX19 | hg38_v1_chr1_+_168280872_168280882 | -0.34 | 9.4e-02 | Click! |
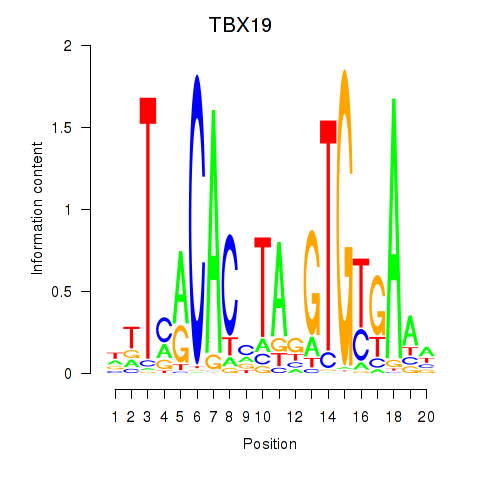
Activity profile of TBX19 motif
Sorted Z-values of TBX19 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX19
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_55160668 | 1.27 |
ENST00000588076.1
|
DNAAF3
|
dynein axonemal assembly factor 3 |
| chr1_-_120051714 | 0.80 |
ENST00000579475.7
|
NOTCH2
|
notch receptor 2 |
| chr15_+_67125707 | 0.68 |
ENST00000540846.6
|
SMAD3
|
SMAD family member 3 |
| chr7_+_101020073 | 0.34 |
ENST00000306151.9
|
MUC17
|
mucin 17, cell surface associated |
| chr5_-_95081482 | 0.32 |
ENST00000312216.12
ENST00000512425.5 ENST00000505208.5 ENST00000429576.6 ENST00000508509.5 ENST00000510732.5 |
MCTP1
|
multiple C2 and transmembrane domain containing 1 |
| chr19_+_37371152 | 0.31 |
ENST00000483919.5
ENST00000588911.5 ENST00000587349.1 |
ZNF527
|
zinc finger protein 527 |
| chr15_-_55289756 | 0.31 |
ENST00000336787.6
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr5_-_178232537 | 0.30 |
ENST00000476170.2
ENST00000323594.8 |
PHYKPL
|
5-phosphohydroxy-L-lysine phospho-lyase |
| chr5_+_55024250 | 0.28 |
ENST00000231009.3
|
GZMK
|
granzyme K |
| chr1_+_46175079 | 0.27 |
ENST00000372003.6
|
TSPAN1
|
tetraspanin 1 |
| chr5_+_96876480 | 0.27 |
ENST00000437043.8
ENST00000379904.8 |
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chr1_+_205257180 | 0.26 |
ENST00000330675.12
|
TMCC2
|
transmembrane and coiled-coil domain family 2 |
| chr4_+_122732652 | 0.25 |
ENST00000542236.5
ENST00000314218.8 |
BBS12
|
Bardet-Biedl syndrome 12 |
| chr5_-_115626161 | 0.25 |
ENST00000282382.8
|
TMED7-TICAM2
|
TMED7-TICAM2 readthrough |
| chr5_+_35856883 | 0.25 |
ENST00000506850.5
ENST00000303115.8 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr3_+_132597260 | 0.24 |
ENST00000249887.3
|
ACKR4
|
atypical chemokine receptor 4 |
| chr20_+_45207025 | 0.24 |
ENST00000372781.4
|
SEMG1
|
semenogelin 1 |
| chr14_-_106154113 | 0.23 |
ENST00000390603.2
|
IGHV3-15
|
immunoglobulin heavy variable 3-15 |
| chr7_+_149147422 | 0.22 |
ENST00000475153.6
|
ZNF398
|
zinc finger protein 398 |
| chr9_-_16727980 | 0.22 |
ENST00000418777.5
ENST00000468187.6 |
BNC2
|
basonuclin 2 |
| chr7_-_139716980 | 0.22 |
ENST00000342645.7
|
HIPK2
|
homeodomain interacting protein kinase 2 |
| chr17_-_41140487 | 0.22 |
ENST00000345847.4
|
KRTAP4-6
|
keratin associated protein 4-6 |
| chr14_+_20781139 | 0.20 |
ENST00000304677.3
|
RNASE6
|
ribonuclease A family member k6 |
| chr7_-_122995700 | 0.20 |
ENST00000249284.3
|
TAS2R16
|
taste 2 receptor member 16 |
| chr19_+_4247074 | 0.20 |
ENST00000262962.12
|
YJU2
|
YJU2 splicing factor homolog |
| chr8_+_13566854 | 0.19 |
ENST00000297324.5
|
C8orf48
|
chromosome 8 open reading frame 48 |
| chr16_+_84768246 | 0.18 |
ENST00000569925.1
ENST00000567526.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chrX_+_76173010 | 0.18 |
ENST00000373357.3
ENST00000373358.8 |
PBDC1
|
polysaccharide biosynthesis domain containing 1 |
| chr3_-_194672175 | 0.18 |
ENST00000265245.10
|
LSG1
|
large 60S subunit nuclear export GTPase 1 |
| chr8_+_32548210 | 0.17 |
ENST00000523079.5
ENST00000650919.1 |
NRG1
|
neuregulin 1 |
| chr2_-_287320 | 0.17 |
ENST00000401503.5
|
ALKAL2
|
ALK and LTK ligand 2 |
| chr2_-_26641363 | 0.17 |
ENST00000288861.5
|
CIB4
|
calcium and integrin binding family member 4 |
| chr12_-_9607903 | 0.17 |
ENST00000229402.4
|
KLRB1
|
killer cell lectin like receptor B1 |
| chr11_-_18939493 | 0.16 |
ENST00000526914.1
|
MRGPRX1
|
MAS related GPR family member X1 |
| chr6_+_10694916 | 0.16 |
ENST00000379568.4
|
PAK1IP1
|
PAK1 interacting protein 1 |
| chr19_+_46229731 | 0.16 |
ENST00000437936.2
|
IGFL1
|
IGF like family member 1 |
| chr1_+_100345018 | 0.16 |
ENST00000635056.2
ENST00000647005.1 |
CDC14A
|
cell division cycle 14A |
| chr7_-_138627444 | 0.15 |
ENST00000463557.1
|
SVOPL
|
SVOP like |
| chr5_-_33984636 | 0.15 |
ENST00000382102.7
ENST00000509381.1 |
SLC45A2
|
solute carrier family 45 member 2 |
| chr6_-_36547400 | 0.15 |
ENST00000229812.8
|
STK38
|
serine/threonine kinase 38 |
| chr3_-_71493500 | 0.15 |
ENST00000648380.1
ENST00000650295.1 |
FOXP1
|
forkhead box P1 |
| chr2_+_143129379 | 0.14 |
ENST00000295095.11
|
ARHGAP15
|
Rho GTPase activating protein 15 |
| chr1_-_193105373 | 0.14 |
ENST00000367439.8
|
GLRX2
|
glutaredoxin 2 |
| chr7_+_66114814 | 0.14 |
ENST00000431089.6
ENST00000398684.6 ENST00000395326.8 ENST00000338592.5 |
CRCP
|
CGRP receptor component |
| chr7_-_151440008 | 0.14 |
ENST00000491928.1
ENST00000644350.1 ENST00000337323.3 |
CRYGN
|
crystallin gamma N |
| chr7_-_38354517 | 0.13 |
ENST00000390345.2
|
TRGV4
|
T cell receptor gamma variable 4 |
| chr1_+_89524819 | 0.13 |
ENST00000439853.6
ENST00000330947.7 ENST00000449440.5 ENST00000640258.1 |
LRRC8B
|
leucine rich repeat containing 8 VRAC subunit B |
| chr4_+_15703057 | 0.13 |
ENST00000265016.9
ENST00000382346.7 |
BST1
|
bone marrow stromal cell antigen 1 |
| chr11_-_22625804 | 0.13 |
ENST00000327470.6
|
FANCF
|
FA complementation group F |
| chr7_-_43906577 | 0.13 |
ENST00000603700.1
ENST00000439702.5 ENST00000453200.6 ENST00000402306.7 ENST00000443736.5 ENST00000446958.1 |
URGCP-MRPS24
URGCP
|
URGCP-MRPS24 readthrough upregulator of cell proliferation |
| chr1_+_171512032 | 0.13 |
ENST00000426496.6
|
PRRC2C
|
proline rich coiled-coil 2C |
| chr16_+_31393329 | 0.12 |
ENST00000389202.3
|
ITGAD
|
integrin subunit alpha D |
| chr14_+_104745960 | 0.12 |
ENST00000556623.1
ENST00000555674.1 |
ADSS1
|
adenylosuccinate synthase 1 |
| chr1_+_89524871 | 0.12 |
ENST00000639264.1
|
LRRC8B
|
leucine rich repeat containing 8 VRAC subunit B |
| chr19_-_7943648 | 0.12 |
ENST00000597926.1
ENST00000270538.8 |
TIMM44
|
translocase of inner mitochondrial membrane 44 |
| chr12_+_52069967 | 0.12 |
ENST00000336854.9
ENST00000550604.1 ENST00000553049.5 ENST00000548915.1 |
ATG101
|
autophagy related 101 |
| chr6_-_127459364 | 0.12 |
ENST00000487331.2
ENST00000483725.8 |
KIAA0408
|
KIAA0408 |
| chr10_-_95441015 | 0.12 |
ENST00000371241.5
ENST00000354106.7 ENST00000371239.5 ENST00000361941.7 ENST00000277982.9 ENST00000371245.7 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr10_+_75210151 | 0.12 |
ENST00000298468.9
ENST00000543351.5 |
VDAC2
|
voltage dependent anion channel 2 |
| chr14_+_55271344 | 0.11 |
ENST00000681400.1
ENST00000679934.1 ENST00000681904.1 ENST00000313833.5 |
FBXO34
|
F-box protein 34 |
| chr7_-_76626127 | 0.11 |
ENST00000454397.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr5_+_161850597 | 0.11 |
ENST00000634335.1
ENST00000635880.1 |
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1 |
| chr22_+_44677044 | 0.11 |
ENST00000006251.11
|
PRR5
|
proline rich 5 |
| chr20_-_3781440 | 0.11 |
ENST00000379756.3
|
SPEF1
|
sperm flagellar 1 |
| chr19_-_7021431 | 0.10 |
ENST00000636986.2
ENST00000637800.1 |
MBD3L2B
|
methyl-CpG binding domain protein 3 like 2B |
| chr2_+_72916183 | 0.10 |
ENST00000394111.6
|
EMX1
|
empty spiracles homeobox 1 |
| chr19_-_2292024 | 0.10 |
ENST00000585527.1
|
LINGO3
|
leucine rich repeat and Ig domain containing 3 |
| chrX_-_19970298 | 0.10 |
ENST00000379687.7
ENST00000379682.8 |
BCLAF3
|
BCLAF1 and THRAP3 family member 3 |
| chr22_+_44677077 | 0.09 |
ENST00000403581.5
|
PRR5
|
proline rich 5 |
| chr3_-_47513677 | 0.09 |
ENST00000296149.9
|
ELP6
|
elongator acetyltransferase complex subunit 6 |
| chr17_-_41047267 | 0.09 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr5_+_96875978 | 0.09 |
ENST00000510373.5
|
ERAP2
|
endoplasmic reticulum aminopeptidase 2 |
| chr10_+_93893931 | 0.09 |
ENST00000371408.7
ENST00000427197.2 |
SLC35G1
|
solute carrier family 35 member G1 |
| chr11_-_59668981 | 0.09 |
ENST00000300146.10
|
PATL1
|
PAT1 homolog 1, processing body mRNA decay factor |
| chr6_-_38639898 | 0.09 |
ENST00000481247.6
|
BTBD9
|
BTB domain containing 9 |
| chr11_+_119121559 | 0.09 |
ENST00000350777.7
ENST00000529988.5 ENST00000527410.3 |
HINFP
|
histone H4 transcription factor |
| chr7_+_112480853 | 0.09 |
ENST00000439068.6
ENST00000312849.4 |
LSMEM1
|
leucine rich single-pass membrane protein 1 |
| chr7_+_117611617 | 0.08 |
ENST00000468795.1
|
CFTR
|
CF transmembrane conductance regulator |
| chr11_+_30323077 | 0.08 |
ENST00000282032.4
|
ARL14EP
|
ADP ribosylation factor like GTPase 14 effector protein |
| chr16_+_83971451 | 0.08 |
ENST00000565691.5
|
NECAB2
|
N-terminal EF-hand calcium binding protein 2 |
| chr19_-_51020019 | 0.08 |
ENST00000309958.7
|
KLK10
|
kallikrein related peptidase 10 |
| chr9_+_75890664 | 0.08 |
ENST00000376767.7
ENST00000674117.1 ENST00000376752.8 |
PCSK5
|
proprotein convertase subtilisin/kexin type 5 |
| chr13_-_96994350 | 0.08 |
ENST00000298440.5
ENST00000543457.6 ENST00000541038.2 |
OXGR1
|
oxoglutarate receptor 1 |
| chr2_+_89851723 | 0.08 |
ENST00000429992.2
|
IGKV2D-40
|
immunoglobulin kappa variable 2D-40 |
| chr3_+_119186716 | 0.08 |
ENST00000460625.1
|
UPK1B
|
uroplakin 1B |
| chr6_-_46735693 | 0.08 |
ENST00000537365.1
|
PLA2G7
|
phospholipase A2 group VII |
| chr1_-_150697128 | 0.08 |
ENST00000427665.1
ENST00000271732.8 |
GOLPH3L
|
golgi phosphoprotein 3 like |
| chr12_+_1970772 | 0.08 |
ENST00000682544.1
|
CACNA1C
|
calcium voltage-gated channel subunit alpha1 C |
| chr20_-_44960348 | 0.08 |
ENST00000372813.4
|
TOMM34
|
translocase of outer mitochondrial membrane 34 |
| chr3_+_46242453 | 0.08 |
ENST00000452454.1
ENST00000395940.3 ENST00000457243.1 |
CCR3
|
C-C motif chemokine receptor 3 |
| chr11_+_57657736 | 0.08 |
ENST00000529773.2
ENST00000533905.1 ENST00000525602.1 ENST00000533682.2 ENST00000302731.4 |
CLP1
|
cleavage factor polyribonucleotide kinase subunit 1 |
| chr12_+_1970809 | 0.08 |
ENST00000683781.1
ENST00000682462.1 |
CACNA1C
|
calcium voltage-gated channel subunit alpha1 C |
| chr6_-_125301900 | 0.08 |
ENST00000608295.5
ENST00000398153.7 ENST00000608284.1 |
HDDC2
|
HD domain containing 2 |
| chr19_+_19528901 | 0.08 |
ENST00000514277.6
|
YJEFN3
|
YjeF N-terminal domain containing 3 |
| chr5_+_140786291 | 0.08 |
ENST00000394633.7
|
PCDHA1
|
protocadherin alpha 1 |
| chr5_-_115625972 | 0.07 |
ENST00000333314.3
ENST00000456936.4 |
TMED7-TICAM2
TMED7
|
TMED7-TICAM2 readthrough transmembrane p24 trafficking protein 7 |
| chr17_-_19745369 | 0.07 |
ENST00000573368.5
ENST00000457500.6 |
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1 |
| chr20_-_22584547 | 0.07 |
ENST00000419308.7
|
FOXA2
|
forkhead box A2 |
| chr17_+_58166982 | 0.07 |
ENST00000545221.2
|
OR4D2
|
olfactory receptor family 4 subfamily D member 2 |
| chr4_-_68951763 | 0.07 |
ENST00000251566.9
|
UGT2A3
|
UDP glucuronosyltransferase family 2 member A3 |
| chr2_+_233917371 | 0.07 |
ENST00000324695.9
ENST00000433712.6 |
TRPM8
|
transient receptor potential cation channel subfamily M member 8 |
| chr8_-_63038788 | 0.07 |
ENST00000518113.2
ENST00000260118.7 ENST00000677482.1 |
GGH
|
gamma-glutamyl hydrolase |
| chr2_-_73642413 | 0.07 |
ENST00000272425.4
|
NAT8
|
N-acetyltransferase 8 (putative) |
| chr6_+_110958680 | 0.07 |
ENST00000329970.8
|
GTF3C6
|
general transcription factor IIIC subunit 6 |
| chr11_-_115504389 | 0.07 |
ENST00000545380.5
ENST00000452722.7 ENST00000331581.11 ENST00000537058.5 ENST00000536727.5 ENST00000542447.6 |
CADM1
|
cell adhesion molecule 1 |
| chr5_+_126360113 | 0.07 |
ENST00000513040.5
|
GRAMD2B
|
GRAM domain containing 2B |
| chr12_-_10453330 | 0.07 |
ENST00000347831.9
ENST00000359151.8 |
KLRC1
|
killer cell lectin like receptor C1 |
| chr19_+_48019726 | 0.07 |
ENST00000593413.1
|
ELSPBP1
|
epididymal sperm binding protein 1 |
| chr19_-_54313074 | 0.07 |
ENST00000486742.2
ENST00000432233.8 |
LILRA5
|
leukocyte immunoglobulin like receptor A5 |
| chr7_+_102748972 | 0.06 |
ENST00000413034.3
ENST00000409231.7 ENST00000418198.5 |
FAM185A
|
family with sequence similarity 185 member A |
| chr11_-_65121780 | 0.06 |
ENST00000525297.5
ENST00000529259.1 |
FAU
|
FAU ubiquitin like and ribosomal protein S30 fusion |
| chrX_+_108045050 | 0.06 |
ENST00000458383.1
ENST00000217957.10 |
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr15_-_68229658 | 0.06 |
ENST00000565471.6
ENST00000637494.1 ENST00000636314.1 ENST00000637667.1 ENST00000564752.1 ENST00000566347.5 ENST00000249806.11 ENST00000562767.2 |
CLN6
ENSG00000260007.3
|
CLN6 transmembrane ER protein novel protein |
| chr3_-_49429252 | 0.06 |
ENST00000615713.4
|
NICN1
|
nicolin 1 |
| chr18_-_21600645 | 0.06 |
ENST00000269214.10
|
ESCO1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr19_-_40090860 | 0.06 |
ENST00000599972.1
ENST00000450241.6 ENST00000595687.6 ENST00000340963.9 |
ZNF780A
|
zinc finger protein 780A |
| chr7_-_138755892 | 0.06 |
ENST00000644341.1
ENST00000478480.2 |
ATP6V0A4
|
ATPase H+ transporting V0 subunit a4 |
| chrX_+_120604084 | 0.06 |
ENST00000371317.10
|
MCTS1
|
MCTS1 re-initiation and release factor |
| chr6_+_42879610 | 0.06 |
ENST00000493763.7
ENST00000304734.9 |
RPL7L1
|
ribosomal protein L7 like 1 |
| chr2_-_213151590 | 0.06 |
ENST00000374319.8
ENST00000457361.5 ENST00000451136.6 ENST00000434687.6 |
IKZF2
|
IKAROS family zinc finger 2 |
| chr7_-_100081758 | 0.06 |
ENST00000424697.5
|
ZNF3
|
zinc finger protein 3 |
| chr5_+_33440696 | 0.06 |
ENST00000502553.5
ENST00000514259.5 |
TARS1
|
threonyl-tRNA synthetase 1 |
| chr19_-_58002761 | 0.06 |
ENST00000552184.1
ENST00000546715.5 ENST00000547828.5 ENST00000547121.5 ENST00000551380.6 |
ZNF606
|
zinc finger protein 606 |
| chr15_+_75347030 | 0.06 |
ENST00000566313.5
ENST00000355059.9 ENST00000568059.1 ENST00000568881.1 |
NEIL1
|
nei like DNA glycosylase 1 |
| chr10_+_5094405 | 0.06 |
ENST00000380554.5
|
AKR1C3
|
aldo-keto reductase family 1 member C3 |
| chr14_+_23988914 | 0.05 |
ENST00000335125.11
|
DHRS4L2
|
dehydrogenase/reductase 4 like 2 |
| chr17_+_50274447 | 0.05 |
ENST00000507382.2
|
TMEM92
|
transmembrane protein 92 |
| chr11_+_61950343 | 0.05 |
ENST00000378043.9
|
BEST1
|
bestrophin 1 |
| chr15_+_41286011 | 0.05 |
ENST00000661438.1
|
ENSG00000285920.2
|
novel protein |
| chr12_-_110689751 | 0.05 |
ENST00000439744.6
ENST00000549442.5 |
HVCN1
|
hydrogen voltage gated channel 1 |
| chr18_-_49849827 | 0.05 |
ENST00000592688.1
|
MYO5B
|
myosin VB |
| chr7_+_2354810 | 0.05 |
ENST00000360876.9
ENST00000413917.5 ENST00000397011.2 |
EIF3B
|
eukaryotic translation initiation factor 3 subunit B |
| chr6_+_26204552 | 0.05 |
ENST00000615164.2
|
H4C5
|
H4 clustered histone 5 |
| chr16_-_55833085 | 0.05 |
ENST00000360526.8
|
CES1
|
carboxylesterase 1 |
| chr18_+_59225492 | 0.05 |
ENST00000456142.3
ENST00000530323.1 |
GRP
|
gastrin releasing peptide |
| chr14_+_69879408 | 0.05 |
ENST00000361956.8
ENST00000381280.4 |
SMOC1
|
SPARC related modular calcium binding 1 |
| chr14_+_23988884 | 0.05 |
ENST00000558753.5
ENST00000537912.5 |
DHRS4L2
|
dehydrogenase/reductase 4 like 2 |
| chr17_+_7219857 | 0.05 |
ENST00000583312.5
ENST00000350303.9 ENST00000584103.5 ENST00000356839.10 ENST00000579886.2 |
ACADVL
|
acyl-CoA dehydrogenase very long chain |
| chr12_-_48925483 | 0.05 |
ENST00000550765.6
ENST00000552878.5 ENST00000453172.2 |
FKBP11
|
FKBP prolyl isomerase 11 |
| chr7_-_148883474 | 0.04 |
ENST00000476773.5
|
EZH2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr6_-_42746054 | 0.04 |
ENST00000372876.2
|
TBCC
|
tubulin folding cofactor C |
| chr9_-_128128102 | 0.04 |
ENST00000617202.4
|
PTGES2
|
prostaglandin E synthase 2 |
| chr20_-_462485 | 0.04 |
ENST00000681414.1
ENST00000680050.1 ENST00000681129.1 ENST00000354200.5 ENST00000679895.1 ENST00000681551.1 ENST00000681539.1 |
TBC1D20
|
TBC1 domain family member 20 |
| chr17_+_36948925 | 0.04 |
ENST00000616434.2
ENST00000680340.1 ENST00000619387.5 ENST00000679997.1 |
AATF
|
apoptosis antagonizing transcription factor |
| chr6_+_22569554 | 0.04 |
ENST00000510882.4
|
HDGFL1
|
HDGF like 1 |
| chr19_+_48993525 | 0.04 |
ENST00000601968.5
ENST00000596837.5 |
RUVBL2
|
RuvB like AAA ATPase 2 |
| chr17_-_19745602 | 0.04 |
ENST00000444455.5
ENST00000439102.6 |
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1 |
| chr20_-_7940444 | 0.04 |
ENST00000378789.4
|
HAO1
|
hydroxyacid oxidase 1 |
| chr3_+_46163465 | 0.04 |
ENST00000357422.2
|
CCR3
|
C-C motif chemokine receptor 3 |
| chr2_-_33599269 | 0.04 |
ENST00000431950.1
ENST00000403368.1 ENST00000238823.13 |
FAM98A
|
family with sequence similarity 98 member A |
| chrX_+_108044967 | 0.04 |
ENST00000415430.7
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr10_-_119536533 | 0.04 |
ENST00000392865.5
|
RGS10
|
regulator of G protein signaling 10 |
| chr8_+_109362453 | 0.04 |
ENST00000378402.10
|
PKHD1L1
|
PKHD1 like 1 |
| chr3_-_142225556 | 0.04 |
ENST00000392993.7
|
GK5
|
glycerol kinase 5 |
| chr4_-_155376876 | 0.04 |
ENST00000433024.5
ENST00000379248.6 |
MAP9
|
microtubule associated protein 9 |
| chr9_-_128127711 | 0.04 |
ENST00000449878.1
ENST00000338961.11 ENST00000678174.1 |
PTGES2
|
prostaglandin E synthase 2 |
| chr16_-_20327801 | 0.04 |
ENST00000381360.9
|
GP2
|
glycoprotein 2 |
| chrX_+_130401962 | 0.04 |
ENST00000305536.11
ENST00000370947.1 |
RBMX2
|
RNA binding motif protein X-linked 2 |
| chr19_+_9247344 | 0.04 |
ENST00000641946.1
|
OR7E24
|
olfactory receptor family 7 subfamily E member 24 |
| chr4_+_70050431 | 0.04 |
ENST00000511674.5
ENST00000246896.8 |
HTN1
|
histatin 1 |
| chr6_-_130956371 | 0.03 |
ENST00000639623.1
ENST00000525193.5 ENST00000527659.5 |
EPB41L2
|
erythrocyte membrane protein band 4.1 like 2 |
| chrX_+_120604199 | 0.03 |
ENST00000371315.3
|
MCTS1
|
MCTS1 re-initiation and release factor |
| chr1_+_116754422 | 0.03 |
ENST00000369478.4
ENST00000369477.1 |
CD2
|
CD2 molecule |
| chr6_-_26216673 | 0.03 |
ENST00000541790.3
|
H2BC8
|
H2B clustered histone 8 |
| chr14_-_106511856 | 0.03 |
ENST00000390622.2
|
IGHV1-46
|
immunoglobulin heavy variable 1-46 |
| chr1_+_113929350 | 0.03 |
ENST00000369559.8
ENST00000626993.2 |
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr20_+_3209469 | 0.03 |
ENST00000380113.8
ENST00000455664.6 ENST00000399838.3 |
ITPA
|
inosine triphosphatase |
| chr1_+_113929600 | 0.03 |
ENST00000369558.5
ENST00000369561.8 |
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr1_-_167935987 | 0.03 |
ENST00000367846.8
|
MPC2
|
mitochondrial pyruvate carrier 2 |
| chr8_+_2045037 | 0.03 |
ENST00000262113.9
|
MYOM2
|
myomesin 2 |
| chr15_+_40351026 | 0.03 |
ENST00000448599.2
|
PHGR1
|
proline, histidine and glycine rich 1 |
| chr3_+_15601969 | 0.03 |
ENST00000436193.5
ENST00000383778.5 |
BTD
|
biotinidase |
| chr11_+_34624244 | 0.03 |
ENST00000529527.5
ENST00000531728.5 ENST00000525253.5 |
EHF
|
ETS homologous factor |
| chr5_-_100903214 | 0.03 |
ENST00000451528.2
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr15_-_93073111 | 0.03 |
ENST00000557420.1
ENST00000542321.6 |
RGMA
|
repulsive guidance molecule BMP co-receptor a |
| chr6_-_137044959 | 0.03 |
ENST00000468393.5
ENST00000367746.3 ENST00000316649.10 |
IL20RA
|
interleukin 20 receptor subunit alpha |
| chr7_-_133082032 | 0.03 |
ENST00000448878.6
ENST00000262570.10 |
CHCHD3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr2_+_90114838 | 0.02 |
ENST00000417279.3
|
IGKV3D-15
|
immunoglobulin kappa variable 3D-15 |
| chr16_+_33827140 | 0.02 |
ENST00000562905.2
|
IGHV3OR16-13
|
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
| chr1_-_100249815 | 0.02 |
ENST00000370131.3
ENST00000681617.1 ENST00000681780.1 ENST00000370132.8 |
DBT
|
dihydrolipoamide branched chain transacylase E2 |
| chr3_-_49429304 | 0.02 |
ENST00000636166.1
ENST00000273598.8 ENST00000436744.2 |
ENSG00000283189.2
NICN1
|
novel protein nicolin 1 |
| chr19_-_40090921 | 0.02 |
ENST00000595508.1
ENST00000414720.6 ENST00000455521.5 ENST00000595773.5 ENST00000683561.1 |
ENSG00000269749.1
ZNF780A
|
novel transcript zinc finger protein 780A |
| chr4_+_26319636 | 0.02 |
ENST00000342295.6
ENST00000506956.5 ENST00000512671.6 ENST00000345843.8 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr12_+_6873561 | 0.02 |
ENST00000433346.5
|
LRRC23
|
leucine rich repeat containing 23 |
| chr8_+_49072335 | 0.02 |
ENST00000399653.8
ENST00000522267.6 ENST00000303202.8 |
PPDPFL
|
pancreatic progenitor cell differentiation and proliferation factor like |
| chr19_+_49487510 | 0.02 |
ENST00000679106.1
ENST00000621674.4 ENST00000391857.9 ENST00000678510.1 ENST00000467825.2 |
RPL13A
|
ribosomal protein L13a |
| chr5_-_78985288 | 0.02 |
ENST00000264914.10
|
ARSB
|
arylsulfatase B |
| chr15_-_93073706 | 0.02 |
ENST00000425933.6
|
RGMA
|
repulsive guidance molecule BMP co-receptor a |
| chr11_+_118527463 | 0.02 |
ENST00000302783.10
|
TTC36
|
tetratricopeptide repeat domain 36 |
| chr9_-_133703700 | 0.02 |
ENST00000371868.5
|
SARDH
|
sarcosine dehydrogenase |
| chr19_-_38847423 | 0.02 |
ENST00000647557.1
|
HNRNPL
|
heterogeneous nuclear ribonucleoprotein L |
| chr1_-_160646958 | 0.02 |
ENST00000538290.2
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1 |
| chr3_+_57756230 | 0.02 |
ENST00000295951.7
ENST00000659705.1 ENST00000671191.1 |
SLMAP
|
sarcolemma associated protein |
| chr2_+_227616998 | 0.02 |
ENST00000641801.1
|
SCYGR4
|
small cysteine and glycine repeat containing 4 |
| chr5_-_160419059 | 0.02 |
ENST00000297151.9
ENST00000519349.5 ENST00000520664.1 |
SLU7
|
SLU7 homolog, splicing factor |
| chr3_-_98035295 | 0.02 |
ENST00000621172.4
|
GABRR3
|
gamma-aminobutyric acid type A receptor subunit rho3 |
| chr6_-_26285526 | 0.02 |
ENST00000377727.2
|
H4C8
|
H4 clustered histone 8 |
| chr2_-_200889266 | 0.02 |
ENST00000443398.5
ENST00000286175.12 ENST00000409449.5 |
PPIL3
|
peptidylprolyl isomerase like 3 |
| chr1_-_160647287 | 0.02 |
ENST00000235739.6
|
SLAMF1
|
signaling lymphocytic activation molecule family member 1 |
| chr5_+_108924585 | 0.02 |
ENST00000618353.1
|
FER
|
FER tyrosine kinase |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 0.3 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.2 | GO:1902573 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.0 | 0.8 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.2 | GO:0070378 | positive regulation of ERK5 cascade(GO:0070378) |
| 0.0 | 0.2 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 1.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.1 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.4 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.3 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:1904021 | negative regulation of G-protein coupled receptor internalization(GO:1904021) |
| 0.0 | 0.1 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.1 | GO:0042791 | tRNA transcription(GO:0009304) 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.0 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.0 | 0.2 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.0 | 0.1 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.0 | 0.2 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.0 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.0 | 0.1 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.2 | GO:0006554 | lysine catabolic process(GO:0006554) |
| 0.0 | 0.0 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.1 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.8 | GO:0038049 | transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) |
| 0.1 | 0.3 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.2 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 0.2 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.2 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.1 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.0 | 0.1 | GO:0052857 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.0 | 0.1 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0047787 | geranylgeranyl reductase activity(GO:0045550) delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.0 | 0.1 | GO:0030613 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.0 | GO:0052853 | (S)-2-hydroxy-acid oxidase activity(GO:0003973) very-long-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052852) long-chain-(S)-2-hydroxy-long-chain-acid oxidase activity(GO:0052853) medium-chain-(S)-2-hydroxy-acid oxidase activity(GO:0052854) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |