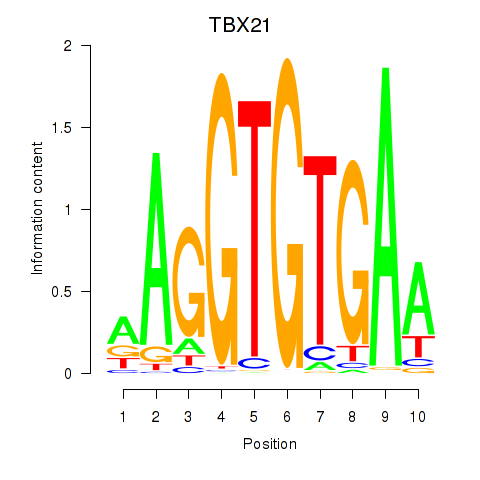
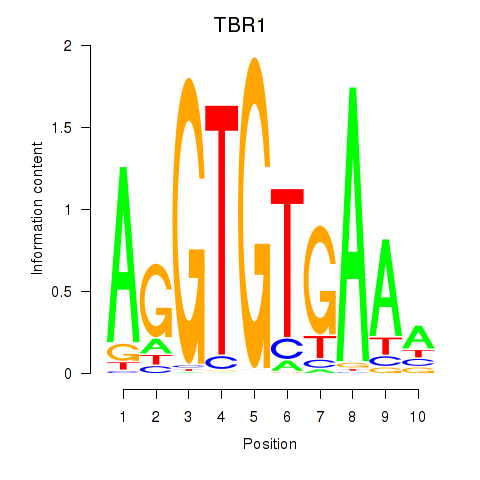
|
chr2_-_156332694
Show fit
|
1.61 |
ENST00000424077.1
ENST00000426264.5
ENST00000421709.1
ENST00000339562.9
|
NR4A2
|
nuclear receptor subfamily 4 group A member 2
|
|
chr1_+_28259473
Show fit
|
0.90 |
ENST00000253063.4
|
SESN2
|
sestrin 2
|
|
chr11_-_27722021
Show fit
|
0.89 |
ENST00000314915.6
|
BDNF
|
brain derived neurotrophic factor
|
|
chr1_+_78620432
Show fit
|
0.81 |
ENST00000370751.10
ENST00000459784.6
ENST00000680110.1
ENST00000680295.1
|
IFI44L
|
interferon induced protein 44 like
|
|
chr4_-_74038681
Show fit
|
0.81 |
ENST00000296026.4
|
CXCL3
|
C-X-C motif chemokine ligand 3
|
|
chr12_+_12785652
Show fit
|
0.74 |
ENST00000356591.5
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr1_+_78620722
Show fit
|
0.74 |
ENST00000679848.1
|
IFI44L
|
interferon induced protein 44 like
|
|
chr19_+_13151975
Show fit
|
0.73 |
ENST00000588173.1
|
IER2
|
immediate early response 2
|
|
chr7_+_27242796
Show fit
|
0.70 |
ENST00000496902.7
|
EVX1
|
even-skipped homeobox 1
|
|
chrX_+_140091415
Show fit
|
0.69 |
ENST00000453380.2
|
ENSG00000230707.2
|
novel protein (LOC389895)
|
|
chr14_+_75278820
Show fit
|
0.66 |
ENST00000554617.1
ENST00000554212.5
ENST00000535987.5
ENST00000303562.9
ENST00000555242.1
|
FOS
|
Fos proto-oncogene, AP-1 transcription factor subunit
|
|
chr22_+_40045451
Show fit
|
0.66 |
ENST00000402203.5
|
TNRC6B
|
trinucleotide repeat containing adaptor 6B
|
|
chr1_-_205321737
Show fit
|
0.61 |
ENST00000367157.6
|
NUAK2
|
NUAK family kinase 2
|
|
chr15_+_60004305
Show fit
|
0.58 |
ENST00000396057.6
|
FOXB1
|
forkhead box B1
|
|
chr6_+_168017873
Show fit
|
0.57 |
ENST00000351261.4
ENST00000354419.6
|
KIF25
|
kinesin family member 25
|
|
chr6_-_13487593
Show fit
|
0.57 |
ENST00000379287.4
ENST00000603223.1
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1
|
|
chr2_+_209771814
Show fit
|
0.56 |
ENST00000673951.1
ENST00000673920.1
|
UNC80
|
unc-80 homolog, NALCN channel complex subunit
|
|
chr6_-_30617232
Show fit
|
0.53 |
ENST00000376511.7
|
PPP1R10
|
protein phosphatase 1 regulatory subunit 10
|
|
chr2_+_209771972
Show fit
|
0.53 |
ENST00000439458.5
ENST00000272845.10
|
UNC80
|
unc-80 homolog, NALCN channel complex subunit
|
|
chr19_-_13102848
Show fit
|
0.51 |
ENST00000264824.5
|
LYL1
|
LYL1 basic helix-loop-helix family member
|
|
chr14_+_75280078
Show fit
|
0.51 |
ENST00000555347.1
|
FOS
|
Fos proto-oncogene, AP-1 transcription factor subunit
|
|
chr1_+_145927105
Show fit
|
0.51 |
ENST00000437797.5
ENST00000601726.3
ENST00000599626.5
ENST00000599147.5
ENST00000595494.5
ENST00000595518.5
ENST00000597144.5
ENST00000599469.5
ENST00000598354.5
ENST00000598103.5
ENST00000600340.5
ENST00000630257.2
ENST00000625258.1
|
LIX1L-AS1
ENSG00000280778.1
|
LIX1L antisense RNA 1
novel protein, lncRNA-POLR3GL readthrough
|
|
chr5_+_98769273
Show fit
|
0.51 |
ENST00000308234.11
|
RGMB
|
repulsive guidance molecule BMP co-receptor b
|
|
chr12_+_130953898
Show fit
|
0.49 |
ENST00000261654.10
|
ADGRD1
|
adhesion G protein-coupled receptor D1
|
|
chr19_-_45245386
Show fit
|
0.49 |
ENST00000413988.3
|
EXOC3L2
|
exocyst complex component 3 like 2
|
|
chr7_-_100168602
Show fit
|
0.48 |
ENST00000423751.2
ENST00000360039.9
|
GAL3ST4
|
galactose-3-O-sulfotransferase 4
|
|
chr21_+_29130630
Show fit
|
0.48 |
ENST00000399926.5
ENST00000399928.6
|
MAP3K7CL
|
MAP3K7 C-terminal like
|
|
chr3_-_39154558
Show fit
|
0.46 |
ENST00000514182.1
|
CSRNP1
|
cysteine and serine rich nuclear protein 1
|
|
chr4_+_73869385
Show fit
|
0.43 |
ENST00000395761.4
|
CXCL1
|
C-X-C motif chemokine ligand 1
|
|
chr21_-_14546351
Show fit
|
0.43 |
ENST00000619120.4
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr5_+_55024250
Show fit
|
0.42 |
ENST00000231009.3
|
GZMK
|
granzyme K
|
|
chr20_-_32657362
Show fit
|
0.42 |
ENST00000360785.6
|
C20orf203
|
chromosome 20 open reading frame 203
|
|
chr4_+_8580387
Show fit
|
0.41 |
ENST00000382487.5
|
GPR78
|
G protein-coupled receptor 78
|
|
chr21_-_14546297
Show fit
|
0.41 |
ENST00000400566.6
ENST00000400564.5
|
SAMSN1
|
SAM domain, SH3 domain and nuclear localization signals 1
|
|
chr17_-_7329266
Show fit
|
0.40 |
ENST00000571887.5
ENST00000315614.11
ENST00000399464.7
ENST00000570460.5
|
NEURL4
|
neuralized E3 ubiquitin protein ligase 4
|
|
chr11_+_64306227
Show fit
|
0.40 |
ENST00000405666.5
ENST00000468670.2
|
ESRRA
|
estrogen related receptor alpha
|
|
chr12_-_90955172
Show fit
|
0.39 |
ENST00000358859.3
|
CCER1
|
coiled-coil glutamate rich protein 1
|
|
chr1_-_163202835
Show fit
|
0.39 |
ENST00000527988.1
ENST00000531476.1
ENST00000530507.5
ENST00000313961.10
|
RGS5
|
regulator of G protein signaling 5
|
|
chr15_+_40569290
Show fit
|
0.39 |
ENST00000315616.12
ENST00000559271.1
ENST00000616318.1
|
RPUSD2
|
RNA pseudouridine synthase domain containing 2
|
|
chr10_+_116545907
Show fit
|
0.38 |
ENST00000369221.2
|
PNLIP
|
pancreatic lipase
|
|
chr4_+_123396785
Show fit
|
0.37 |
ENST00000505319.5
ENST00000651917.1
ENST00000610581.4
ENST00000339241.1
|
SPRY1
|
sprouty RTK signaling antagonist 1
|
|
chr21_-_30497160
Show fit
|
0.36 |
ENST00000334058.3
|
KRTAP19-4
|
keratin associated protein 19-4
|
|
chr12_-_7872902
Show fit
|
0.36 |
ENST00000431042.7
|
SLC2A14
|
solute carrier family 2 member 14
|
|
chr3_-_61251376
Show fit
|
0.36 |
ENST00000476844.5
ENST00000488467.5
ENST00000492590.6
ENST00000468189.5
|
FHIT
|
fragile histidine triad diadenosine triphosphatase
|
|
chr5_-_35938572
Show fit
|
0.35 |
ENST00000651391.1
ENST00000397366.5
ENST00000513623.5
ENST00000514524.2
ENST00000397367.6
|
CAPSL
|
calcyphosine like
|
|
chr10_+_92691813
Show fit
|
0.34 |
ENST00000472590.6
|
HHEX
|
hematopoietically expressed homeobox
|
|
chr1_+_182789765
Show fit
|
0.34 |
ENST00000367555.5
ENST00000367553.6
|
NPL
|
N-acetylneuraminate pyruvate lyase
|
|
chr11_+_112961480
Show fit
|
0.33 |
ENST00000621850.4
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr17_+_27471999
Show fit
|
0.32 |
ENST00000583370.5
ENST00000509603.6
ENST00000268763.10
ENST00000398988.7
|
KSR1
|
kinase suppressor of ras 1
|
|
chr17_+_42798779
Show fit
|
0.32 |
ENST00000585355.5
|
CNTD1
|
cyclin N-terminal domain containing 1
|
|
chrX_-_129654946
Show fit
|
0.32 |
ENST00000429967.3
|
APLN
|
apelin
|
|
chr5_+_134525649
Show fit
|
0.32 |
ENST00000282605.8
ENST00000681547.2
ENST00000361895.6
ENST00000402835.5
|
JADE2
|
jade family PHD finger 2
|
|
chr3_-_129688691
Show fit
|
0.31 |
ENST00000432054.6
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr5_+_150660841
Show fit
|
0.30 |
ENST00000297130.4
|
MYOZ3
|
myozenin 3
|
|
chr19_+_38289138
Show fit
|
0.30 |
ENST00000590738.1
ENST00000587519.4
ENST00000591889.2
|
SPINT2
ENSG00000267748.4
|
serine peptidase inhibitor, Kunitz type 2
novel protein
|
|
chr12_-_7872802
Show fit
|
0.29 |
ENST00000535344.5
ENST00000543909.5
|
SLC2A14
|
solute carrier family 2 member 14
|
|
chr15_+_89776326
Show fit
|
0.28 |
ENST00000341735.5
|
MESP2
|
mesoderm posterior bHLH transcription factor 2
|
|
chr12_-_7872843
Show fit
|
0.28 |
ENST00000340749.9
ENST00000535295.5
ENST00000539234.5
|
SLC2A14
|
solute carrier family 2 member 14
|
|
chr7_-_27095972
Show fit
|
0.28 |
ENST00000355633.5
ENST00000643460.2
|
HOXA1
|
homeobox A1
|
|
chr11_-_111879425
Show fit
|
0.27 |
ENST00000622211.4
|
ENSG00000258529.5
|
novel protein
|
|
chr20_+_31605280
Show fit
|
0.27 |
ENST00000376105.4
ENST00000376112.4
|
ID1
|
inhibitor of DNA binding 1, HLH protein
|
|
chr2_+_143129379
Show fit
|
0.27 |
ENST00000295095.11
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr16_+_30201057
Show fit
|
0.26 |
ENST00000569485.5
|
SULT1A3
|
sulfotransferase family 1A member 3
|
|
chr1_+_147541491
Show fit
|
0.26 |
ENST00000683836.1
ENST00000234739.8
|
BCL9
|
BCL9 transcription coactivator
|
|
chr7_-_36724380
Show fit
|
0.25 |
ENST00000617267.4
|
AOAH
|
acyloxyacyl hydrolase
|
|
chr12_-_52367478
Show fit
|
0.25 |
ENST00000257901.7
|
KRT85
|
keratin 85
|
|
chr5_+_75511756
Show fit
|
0.25 |
ENST00000241436.8
|
POLK
|
DNA polymerase kappa
|
|
chr11_-_28108109
Show fit
|
0.25 |
ENST00000263181.7
|
KIF18A
|
kinesin family member 18A
|
|
chr16_+_69106219
Show fit
|
0.25 |
ENST00000219322.7
|
HAS3
|
hyaluronan synthase 3
|
|
chr5_+_35856883
Show fit
|
0.24 |
ENST00000506850.5
ENST00000303115.8
ENST00000511982.1
|
IL7R
|
interleukin 7 receptor
|
|
chr22_-_17221841
Show fit
|
0.24 |
ENST00000449907.7
ENST00000441548.1
ENST00000399839.5
|
ADA2
|
adenosine deaminase 2
|
|
chr5_+_176238365
Show fit
|
0.24 |
ENST00000341199.10
ENST00000430704.6
ENST00000443967.5
ENST00000429602.7
|
SIMC1
|
SUMO interacting motifs containing 1
|
|
chr16_-_20669855
Show fit
|
0.23 |
ENST00000524149.5
|
ACSM1
|
acyl-CoA synthetase medium chain family member 1
|
|
chr5_+_150661243
Show fit
|
0.23 |
ENST00000517768.6
|
MYOZ3
|
myozenin 3
|
|
chr1_-_154859841
Show fit
|
0.23 |
ENST00000361147.8
|
KCNN3
|
potassium calcium-activated channel subfamily N member 3
|
|
chr7_+_100429823
Show fit
|
0.23 |
ENST00000310512.4
|
MEPCE
|
methylphosphate capping enzyme
|
|
chr10_-_22714531
Show fit
|
0.22 |
ENST00000376573.9
|
PIP4K2A
|
phosphatidylinositol-5-phosphate 4-kinase type 2 alpha
|
|
chr7_-_36724543
Show fit
|
0.22 |
ENST00000612871.4
|
AOAH
|
acyloxyacyl hydrolase
|
|
chr6_-_22302826
Show fit
|
0.22 |
ENST00000651245.1
|
PRL
|
prolactin
|
|
chr16_+_11965193
Show fit
|
0.22 |
ENST00000053243.6
ENST00000396495.3
|
TNFRSF17
|
TNF receptor superfamily member 17
|
|
chr15_+_47184076
Show fit
|
0.22 |
ENST00000558014.5
|
SEMA6D
|
semaphorin 6D
|
|
chr7_+_143316105
Show fit
|
0.22 |
ENST00000343257.7
ENST00000650516.1
|
CLCN1
|
chloride voltage-gated channel 1
|
|
chr17_-_8163522
Show fit
|
0.22 |
ENST00000404970.3
|
VAMP2
|
vesicle associated membrane protein 2
|
|
chr12_-_52703522
Show fit
|
0.22 |
ENST00000341809.8
|
KRT77
|
keratin 77
|
|
chr20_-_1557679
Show fit
|
0.21 |
ENST00000381621.5
ENST00000381623.4
|
SIRPD
|
signal regulatory protein delta
|
|
chr19_-_43198079
Show fit
|
0.21 |
ENST00000597374.5
ENST00000599371.1
|
PSG4
|
pregnancy specific beta-1-glycoprotein 4
|
|
chr12_+_109052564
Show fit
|
0.21 |
ENST00000257548.10
ENST00000536723.5
ENST00000536393.5
|
USP30
|
ubiquitin specific peptidase 30
|
|
chr1_-_26306576
Show fit
|
0.21 |
ENST00000421827.2
ENST00000374215.5
ENST00000374223.5
ENST00000357089.8
ENST00000314675.11
ENST00000423664.5
ENST00000374221.7
|
UBXN11
|
UBX domain protein 11
|
|
chr17_-_56833889
Show fit
|
0.20 |
ENST00000397861.7
|
C17orf67
|
chromosome 17 open reading frame 67
|
|
chr11_+_73950985
Show fit
|
0.20 |
ENST00000339764.6
|
DNAJB13
|
DnaJ heat shock protein family (Hsp40) member B13
|
|
chr1_+_172659095
Show fit
|
0.20 |
ENST00000367721.3
ENST00000340030.4
|
FASLG
|
Fas ligand
|
|
chr19_+_48019726
Show fit
|
0.20 |
ENST00000593413.1
|
ELSPBP1
|
epididymal sperm binding protein 1
|
|
chr10_+_92691897
Show fit
|
0.20 |
ENST00000492654.3
|
HHEX
|
hematopoietically expressed homeobox
|
|
chr17_+_44847874
Show fit
|
0.20 |
ENST00000253410.3
|
HIGD1B
|
HIG1 hypoxia inducible domain family member 1B
|
|
chrX_-_19670983
Show fit
|
0.20 |
ENST00000379716.5
|
SH3KBP1
|
SH3 domain containing kinase binding protein 1
|
|
chr4_+_185209577
Show fit
|
0.20 |
ENST00000652585.1
|
SNX25
|
sorting nexin 25
|
|
chr1_-_51990679
Show fit
|
0.20 |
ENST00000371655.4
|
RAB3B
|
RAB3B, member RAS oncogene family
|
|
chr16_-_4345904
Show fit
|
0.19 |
ENST00000571941.5
|
PAM16
|
presequence translocase associated motor 16
|
|
chr7_-_36724457
Show fit
|
0.19 |
ENST00000617537.5
ENST00000435386.1
|
AOAH
|
acyloxyacyl hydrolase
|
|
chr15_+_63504511
Show fit
|
0.19 |
ENST00000540797.5
|
USP3
|
ubiquitin specific peptidase 3
|
|
chr15_-_82647336
Show fit
|
0.19 |
ENST00000617522.4
ENST00000684509.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr17_+_36103819
Show fit
|
0.19 |
ENST00000615863.2
ENST00000621626.1
|
CCL4
|
C-C motif chemokine ligand 4
|
|
chr2_+_232456146
Show fit
|
0.19 |
ENST00000295463.4
|
ALPI
|
alkaline phosphatase, intestinal
|
|
chr22_+_18110305
Show fit
|
0.19 |
ENST00000680175.1
ENST00000426208.5
|
TUBA8
|
tubulin alpha 8
|
|
chrX_-_111410420
Show fit
|
0.18 |
ENST00000371993.7
ENST00000680476.1
|
DCX
|
doublecortin
|
|
chr14_+_77116562
Show fit
|
0.18 |
ENST00000557408.5
|
TMEM63C
|
transmembrane protein 63C
|
|
chr17_+_44847905
Show fit
|
0.18 |
ENST00000587021.1
|
HIGD1B
|
HIG1 hypoxia inducible domain family member 1B
|
|
chr2_+_153871909
Show fit
|
0.18 |
ENST00000392825.8
ENST00000434213.1
|
GALNT13
|
polypeptide N-acetylgalactosaminyltransferase 13
|
|
chr15_+_63504583
Show fit
|
0.18 |
ENST00000380324.8
ENST00000561442.5
ENST00000560070.5
|
USP3
|
ubiquitin specific peptidase 3
|
|
chr16_-_30534819
Show fit
|
0.17 |
ENST00000395094.3
|
ZNF747
|
zinc finger protein 747
|
|
chr2_-_218166951
Show fit
|
0.17 |
ENST00000295683.3
|
CXCR1
|
C-X-C motif chemokine receptor 1
|
|
chr2_-_150487658
Show fit
|
0.17 |
ENST00000375734.6
ENST00000263895.9
ENST00000454202.5
|
RND3
|
Rho family GTPase 3
|
|
chr2_+_218382265
Show fit
|
0.17 |
ENST00000233202.11
|
SLC11A1
|
solute carrier family 11 member 1
|
|
chr22_-_40819324
Show fit
|
0.17 |
ENST00000435456.7
ENST00000434185.1
ENST00000544408.5
|
SLC25A17
|
solute carrier family 25 member 17
|
|
chr14_+_20999255
Show fit
|
0.17 |
ENST00000554422.5
ENST00000298681.5
|
SLC39A2
|
solute carrier family 39 member 2
|
|
chr17_+_42798881
Show fit
|
0.17 |
ENST00000588527.5
|
CNTD1
|
cyclin N-terminal domain containing 1
|
|
chr17_-_61928005
Show fit
|
0.17 |
ENST00000444766.7
|
INTS2
|
integrator complex subunit 2
|
|
chr17_+_42798857
Show fit
|
0.17 |
ENST00000588408.6
|
CNTD1
|
cyclin N-terminal domain containing 1
|
|
chr17_-_61927968
Show fit
|
0.17 |
ENST00000646954.1
ENST00000251334.7
ENST00000647009.1
|
INTS2
|
integrator complex subunit 2
|
|
chr17_-_2711633
Show fit
|
0.16 |
ENST00000435359.5
|
CLUH
|
clustered mitochondria homolog
|
|
chr11_+_62665305
Show fit
|
0.16 |
ENST00000532971.2
|
CSKMT
|
citrate synthase lysine methyltransferase
|
|
chr9_+_136658854
Show fit
|
0.16 |
ENST00000371699.5
|
EGFL7
|
EGF like domain multiple 7
|
|
chr8_-_92095215
Show fit
|
0.16 |
ENST00000360348.6
ENST00000520428.5
ENST00000518992.5
ENST00000520556.5
ENST00000518317.5
ENST00000521319.5
ENST00000521375.5
ENST00000518449.5
ENST00000613886.4
|
RUNX1T1
|
RUNX1 partner transcriptional co-repressor 1
|
|
chr8_-_98942557
Show fit
|
0.16 |
ENST00000523601.5
|
STK3
|
serine/threonine kinase 3
|
|
chr7_-_44140372
Show fit
|
0.16 |
ENST00000446581.5
|
MYL7
|
myosin light chain 7
|
|
chr1_+_184386978
Show fit
|
0.16 |
ENST00000235307.7
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr12_+_57460127
Show fit
|
0.16 |
ENST00000532291.5
ENST00000543426.5
ENST00000546141.5
|
GLI1
|
GLI family zinc finger 1
|
|
chr6_-_32192845
Show fit
|
0.16 |
ENST00000487761.5
|
GPSM3
|
G protein signaling modulator 3
|
|
chr6_+_167325071
Show fit
|
0.16 |
ENST00000649884.1
ENST00000239587.10
|
TTLL2
|
tubulin tyrosine ligase like 2
|
|
chr11_+_112961402
Show fit
|
0.16 |
ENST00000613217.4
ENST00000316851.12
ENST00000620046.4
ENST00000531044.5
ENST00000529356.5
|
NCAM1
|
neural cell adhesion molecule 1
|
|
chr14_+_23258485
Show fit
|
0.16 |
ENST00000399905.5
ENST00000470456.1
|
RNF212B
|
ring finger protein 212B
|
|
chr12_+_57459782
Show fit
|
0.16 |
ENST00000228682.7
|
GLI1
|
GLI family zinc finger 1
|
|
chr14_-_75980993
Show fit
|
0.15 |
ENST00000556285.1
|
TGFB3
|
transforming growth factor beta 3
|
|
chr2_+_241188509
Show fit
|
0.15 |
ENST00000674324.1
ENST00000274979.12
|
ANO7
|
anoctamin 7
|
|
chr5_-_138178599
Show fit
|
0.15 |
ENST00000454473.5
ENST00000418329.5
ENST00000254900.10
ENST00000230901.9
ENST00000402931.5
ENST00000411594.6
ENST00000430331.1
|
BRD8
|
bromodomain containing 8
|
|
chr7_+_27242700
Show fit
|
0.15 |
ENST00000222761.7
|
EVX1
|
even-skipped homeobox 1
|
|
chr1_-_11847772
Show fit
|
0.15 |
ENST00000376480.7
ENST00000610706.1
|
NPPA
|
natriuretic peptide A
|
|
chr6_+_30345131
Show fit
|
0.15 |
ENST00000433076.6
ENST00000442966.7
ENST00000428040.6
ENST00000436442.2
|
RPP21
|
ribonuclease P/MRP subunit p21
|
|
chr12_-_52291515
Show fit
|
0.15 |
ENST00000615839.1
ENST00000327741.9
|
KRT81
|
keratin 81
|
|
chr3_-_100993448
Show fit
|
0.15 |
ENST00000495063.6
ENST00000486770.7
ENST00000530539.2
|
ABI3BP
|
ABI family member 3 binding protein
|
|
chr1_+_150364136
Show fit
|
0.15 |
ENST00000369068.5
|
RPRD2
|
regulation of nuclear pre-mRNA domain containing 2
|
|
chr11_+_28108248
Show fit
|
0.15 |
ENST00000406787.7
ENST00000403099.5
ENST00000407364.8
|
METTL15
|
methyltransferase like 15
|
|
chr17_-_41149823
Show fit
|
0.15 |
ENST00000343246.6
|
KRTAP4-5
|
keratin associated protein 4-5
|
|
chr20_-_45515612
Show fit
|
0.15 |
ENST00000217428.7
|
SPINT3
|
serine peptidase inhibitor, Kunitz type 3
|
|
chr12_-_76486061
Show fit
|
0.15 |
ENST00000548341.5
|
OSBPL8
|
oxysterol binding protein like 8
|
|
chr19_+_12751789
Show fit
|
0.14 |
ENST00000553030.6
|
BEST2
|
bestrophin 2
|
|
chr10_+_7818497
Show fit
|
0.14 |
ENST00000344293.6
|
TAF3
|
TATA-box binding protein associated factor 3
|
|
chrX_+_110002635
Show fit
|
0.14 |
ENST00000372072.7
|
TMEM164
|
transmembrane protein 164
|
|
chr15_-_43266857
Show fit
|
0.14 |
ENST00000349114.8
ENST00000220420.10
|
TGM5
|
transglutaminase 5
|
|
chr20_-_290364
Show fit
|
0.14 |
ENST00000382369.9
|
C20orf96
|
chromosome 20 open reading frame 96
|
|
chr7_+_69967464
Show fit
|
0.14 |
ENST00000664521.1
|
AUTS2
|
activator of transcription and developmental regulator AUTS2
|
|
chr19_+_45864318
Show fit
|
0.14 |
ENST00000302177.3
|
FOXA3
|
forkhead box A3
|
|
chr1_+_200027605
Show fit
|
0.14 |
ENST00000236914.7
|
NR5A2
|
nuclear receptor subfamily 5 group A member 2
|
|
chr9_-_136746006
Show fit
|
0.14 |
ENST00000476567.1
|
LCN6
|
lipocalin 6
|
|
chr8_-_42768781
Show fit
|
0.14 |
ENST00000276410.7
|
CHRNA6
|
cholinergic receptor nicotinic alpha 6 subunit
|
|
chr8_-_42768602
Show fit
|
0.13 |
ENST00000534622.5
|
CHRNA6
|
cholinergic receptor nicotinic alpha 6 subunit
|
|
chr14_+_63761836
Show fit
|
0.13 |
ENST00000674003.1
|
SYNE2
|
spectrin repeat containing nuclear envelope protein 2
|
|
chr5_-_71067601
Show fit
|
0.13 |
ENST00000274400.9
ENST00000521602.2
ENST00000330280.11
ENST00000517900.5
|
GTF2H2
|
general transcription factor IIH subunit 2
|
|
chr2_+_170715317
Show fit
|
0.13 |
ENST00000375281.4
|
SP5
|
Sp5 transcription factor
|
|
chr15_-_72231583
Show fit
|
0.13 |
ENST00000566809.1
ENST00000567087.5
ENST00000569050.1
ENST00000568883.5
|
PKM
|
pyruvate kinase M1/2
|
|
chr11_-_236326
Show fit
|
0.13 |
ENST00000525237.1
ENST00000382743.9
ENST00000532956.5
ENST00000525319.5
ENST00000524564.5
|
SIRT3
|
sirtuin 3
|
|
chr1_-_11848345
Show fit
|
0.13 |
ENST00000376476.1
|
NPPA
|
natriuretic peptide A
|
|
chr9_+_122519141
Show fit
|
0.13 |
ENST00000340750.1
|
OR1J4
|
olfactory receptor family 1 subfamily J member 4
|
|
chr1_-_120176450
Show fit
|
0.13 |
ENST00000578049.4
|
SEC22B
|
SEC22 homolog B, vesicle trafficking protein
|
|
chr5_-_75511596
Show fit
|
0.13 |
ENST00000643158.1
ENST00000261415.12
ENST00000646713.1
ENST00000643773.1
ENST00000645866.1
ENST00000644072.2
ENST00000643780.2
ENST00000645483.1
ENST00000642556.1
ENST00000646511.1
|
CERT1
|
ceramide transporter 1
|
|
chr6_+_36678699
Show fit
|
0.13 |
ENST00000405375.5
ENST00000244741.10
ENST00000373711.3
|
CDKN1A
|
cyclin dependent kinase inhibitor 1A
|
|
chr1_+_15736251
Show fit
|
0.13 |
ENST00000294454.6
|
SLC25A34
|
solute carrier family 25 member 34
|
|
chr1_+_26787926
Show fit
|
0.13 |
ENST00000674202.1
ENST00000674222.1
|
PIGV
|
phosphatidylinositol glycan anchor biosynthesis class V
|
|
chr19_+_12751672
Show fit
|
0.13 |
ENST00000549706.5
|
BEST2
|
bestrophin 2
|
|
chr17_+_6756035
Show fit
|
0.12 |
ENST00000361842.8
ENST00000574907.5
|
XAF1
|
XIAP associated factor 1
|
|
chr11_+_119149029
Show fit
|
0.12 |
ENST00000619701.5
|
ABCG4
|
ATP binding cassette subfamily G member 4
|
|
chr6_-_52840843
Show fit
|
0.12 |
ENST00000370989.6
|
GSTA5
|
glutathione S-transferase alpha 5
|
|
chr12_+_64780465
Show fit
|
0.12 |
ENST00000542120.6
|
TBC1D30
|
TBC1 domain family member 30
|
|
chr20_-_38033424
Show fit
|
0.12 |
ENST00000373447.8
ENST00000373448.6
|
TTI1
|
TELO2 interacting protein 1
|
|
chr6_+_43182165
Show fit
|
0.12 |
ENST00000372647.6
ENST00000252050.9
|
CUL9
|
cullin 9
|
|
chr11_+_6239068
Show fit
|
0.12 |
ENST00000379936.3
|
CNGA4
|
cyclic nucleotide gated channel subunit alpha 4
|
|
chr16_-_75248190
Show fit
|
0.12 |
ENST00000542031.6
|
BCAR1
|
BCAR1 scaffold protein, Cas family member
|
|
chr7_-_20786879
Show fit
|
0.12 |
ENST00000418710.3
ENST00000617581.4
ENST00000361443.4
|
SP8
|
Sp8 transcription factor
|
|
chr6_-_84227596
Show fit
|
0.12 |
ENST00000257766.8
|
CEP162
|
centrosomal protein 162
|
|
chrX_-_101291325
Show fit
|
0.12 |
ENST00000356784.2
|
TAF7L
|
TATA-box binding protein associated factor 7 like
|
|
chr19_-_344786
Show fit
|
0.11 |
ENST00000264819.7
|
MIER2
|
MIER family member 2
|
|
chr2_-_85668172
Show fit
|
0.11 |
ENST00000428225.5
ENST00000519937.7
|
SFTPB
|
surfactant protein B
|
|
chr6_-_84227634
Show fit
|
0.11 |
ENST00000617909.1
ENST00000403245.8
|
CEP162
|
centrosomal protein 162
|
|
chr7_+_111091006
Show fit
|
0.11 |
ENST00000451085.5
ENST00000422987.3
ENST00000421101.1
|
LRRN3
|
leucine rich repeat neuronal 3
|
|
chr7_-_138701352
Show fit
|
0.11 |
ENST00000421622.5
ENST00000674285.1
|
SVOPL
|
SVOP like
|
|
chr22_+_30607145
Show fit
|
0.11 |
ENST00000405742.7
|
TCN2
|
transcobalamin 2
|
|
chr16_+_88706458
Show fit
|
0.11 |
ENST00000312060.9
ENST00000453996.7
ENST00000567949.5
ENST00000564921.1
|
CTU2
|
cytosolic thiouridylase subunit 2
|
|
chr17_-_10026265
Show fit
|
0.11 |
ENST00000437099.6
ENST00000396115.6
|
GAS7
|
growth arrest specific 7
|
|
chr2_+_60881553
Show fit
|
0.11 |
ENST00000394479.4
|
REL
|
REL proto-oncogene, NF-kB subunit
|
|
chrX_+_152759218
Show fit
|
0.11 |
ENST00000599845.3
ENST00000638835.1
|
CSAG3
|
CSAG family member 3
|
|
chr8_+_22570944
Show fit
|
0.11 |
ENST00000517962.1
|
SORBS3
|
sorbin and SH3 domain containing 3
|
|
chr20_-_290717
Show fit
|
0.11 |
ENST00000360321.7
ENST00000400269.4
|
C20orf96
|
chromosome 20 open reading frame 96
|
|
chr19_-_50025936
Show fit
|
0.11 |
ENST00000596445.5
ENST00000599538.5
|
VRK3
|
VRK serine/threonine kinase 3
|
|
chr19_+_37371152
Show fit
|
0.11 |
ENST00000483919.5
ENST00000588911.5
ENST00000587349.1
|
ZNF527
|
zinc finger protein 527
|
|
chr2_+_1414382
Show fit
|
0.11 |
ENST00000423320.5
ENST00000346956.7
ENST00000382198.5
|
TPO
|
thyroid peroxidase
|
|
chr17_-_39401593
Show fit
|
0.11 |
ENST00000394294.7
ENST00000264658.11
ENST00000583610.5
ENST00000647139.1
|
FBXL20
|
F-box and leucine rich repeat protein 20
|
|
chr15_-_89751292
Show fit
|
0.11 |
ENST00000300057.4
|
MESP1
|
mesoderm posterior bHLH transcription factor 1
|
|
chr2_+_60881515
Show fit
|
0.11 |
ENST00000295025.12
|
REL
|
REL proto-oncogene, NF-kB subunit
|
|
chr16_+_6019071
Show fit
|
0.11 |
ENST00000547605.5
ENST00000553186.5
|
RBFOX1
|
RNA binding fox-1 homolog 1
|
|
chr10_+_112375196
Show fit
|
0.11 |
ENST00000393081.6
|
ACSL5
|
acyl-CoA synthetase long chain family member 5
|