Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
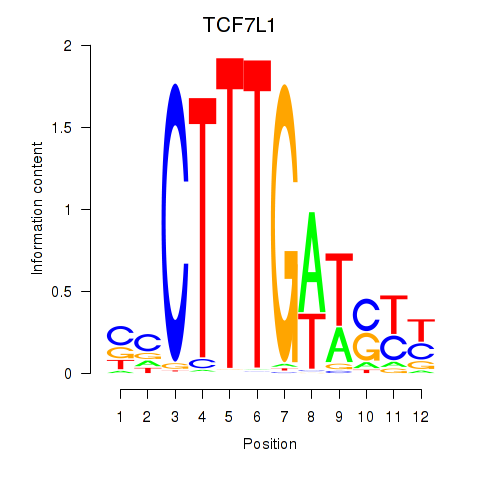
Results for TCF7L1
Z-value: 0.73
Transcription factors associated with TCF7L1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TCF7L1
|
ENSG00000152284.5 | TCF7L1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TCF7L1 | hg38_v1_chr2_+_85133376_85133474 | 0.15 | 4.8e-01 | Click! |
Activity profile of TCF7L1 motif
Sorted Z-values of TCF7L1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TCF7L1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_71867192 | 1.72 |
ENST00000611519.4
ENST00000620444.4 ENST00000613252.5 |
DACH1
|
dachshund family transcription factor 1 |
| chr4_-_185956348 | 1.48 |
ENST00000431902.5
ENST00000284776.11 ENST00000415274.5 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_+_99646025 | 1.29 |
ENST00000263174.9
ENST00000605497.5 ENST00000615664.1 |
PALMD
|
palmdelphin |
| chr1_+_60865259 | 1.19 |
ENST00000371191.5
|
NFIA
|
nuclear factor I A |
| chr2_-_207166818 | 0.94 |
ENST00000423015.5
|
KLF7
|
Kruppel like factor 7 |
| chr16_-_46831043 | 0.91 |
ENST00000565112.1
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr2_+_69013170 | 0.85 |
ENST00000303714.9
|
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr2_+_227813834 | 0.74 |
ENST00000358813.5
ENST00000409189.7 |
CCL20
|
C-C motif chemokine ligand 20 |
| chr14_+_23377136 | 0.67 |
ENST00000382809.2
|
CMTM5
|
CKLF like MARVEL transmembrane domain containing 5 |
| chr2_-_207167220 | 0.66 |
ENST00000421199.5
ENST00000457962.5 |
KLF7
|
Kruppel like factor 7 |
| chrX_+_72069659 | 0.62 |
ENST00000631375.1
|
NHSL2
|
NHS like 2 |
| chr5_+_175861628 | 0.62 |
ENST00000509837.5
|
CPLX2
|
complexin 2 |
| chr10_+_123135938 | 0.60 |
ENST00000357878.7
|
HMX3
|
H6 family homeobox 3 |
| chr3_-_15797930 | 0.60 |
ENST00000683139.1
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr3_-_65622073 | 0.58 |
ENST00000621418.4
ENST00000611645.4 |
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr9_+_125748175 | 0.57 |
ENST00000491787.7
ENST00000447726.6 |
PBX3
|
PBX homeobox 3 |
| chr2_+_11539833 | 0.57 |
ENST00000263834.9
|
GREB1
|
growth regulating estrogen receptor binding 1 |
| chr15_+_62066975 | 0.56 |
ENST00000355522.5
|
C2CD4A
|
C2 calcium dependent domain containing 4A |
| chrX_-_40097403 | 0.55 |
ENST00000397354.7
|
BCOR
|
BCL6 corepressor |
| chr20_+_57391388 | 0.54 |
ENST00000356208.10
ENST00000440234.6 |
RBM38
|
RNA binding motif protein 38 |
| chr14_+_20768393 | 0.53 |
ENST00000326783.4
|
EDDM3B
|
epididymal protein 3B |
| chr10_+_69802424 | 0.52 |
ENST00000673802.2
ENST00000517713.5 ENST00000520133.5 ENST00000522165.5 ENST00000673641.2 ENST00000673628.2 |
COL13A1
|
collagen type XIII alpha 1 chain |
| chr5_+_171419635 | 0.51 |
ENST00000274625.6
|
FGF18
|
fibroblast growth factor 18 |
| chr2_+_69013282 | 0.48 |
ENST00000409829.7
|
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr10_-_86957582 | 0.46 |
ENST00000372027.10
|
MMRN2
|
multimerin 2 |
| chr10_+_69801874 | 0.46 |
ENST00000357811.8
|
COL13A1
|
collagen type XIII alpha 1 chain |
| chr20_+_32010429 | 0.45 |
ENST00000452892.3
ENST00000262659.12 |
CCM2L
|
CCM2 like scaffold protein |
| chr14_+_74763308 | 0.45 |
ENST00000325680.12
ENST00000552421.5 |
YLPM1
|
YLP motif containing 1 |
| chr6_-_11778781 | 0.44 |
ENST00000414691.8
ENST00000229583.9 |
ADTRP
|
androgen dependent TFPI regulating protein |
| chr15_+_81000913 | 0.43 |
ENST00000267984.4
|
TLNRD1
|
talin rod domain containing 1 |
| chr13_+_31945826 | 0.42 |
ENST00000647500.1
|
FRY
|
FRY microtubule binding protein |
| chr10_+_52314272 | 0.42 |
ENST00000373970.4
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr1_-_93614091 | 0.41 |
ENST00000370247.7
|
BCAR3
|
BCAR3 adaptor protein, NSP family member |
| chr11_-_62707413 | 0.41 |
ENST00000360796.10
ENST00000449636.6 |
BSCL2
|
BSCL2 lipid droplet biogenesis associated, seipin |
| chr2_+_69013379 | 0.41 |
ENST00000409349.7
|
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr17_+_82458174 | 0.40 |
ENST00000579198.5
ENST00000390006.8 ENST00000580296.5 |
NARF
|
nuclear prelamin A recognition factor |
| chr2_+_69013414 | 0.39 |
ENST00000681816.1
ENST00000482235.2 |
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr2_+_15940537 | 0.39 |
ENST00000281043.4
ENST00000638417.1 |
MYCN
|
MYCN proto-oncogene, bHLH transcription factor |
| chr10_+_69801892 | 0.39 |
ENST00000398978.8
ENST00000645393.2 ENST00000354547.7 ENST00000674121.1 ENST00000673842.1 ENST00000520267.5 |
COL13A1
|
collagen type XIII alpha 1 chain |
| chr1_+_100351698 | 0.38 |
ENST00000644676.1
|
CDC14A
|
cell division cycle 14A |
| chr10_+_113553039 | 0.38 |
ENST00000351270.4
|
HABP2
|
hyaluronan binding protein 2 |
| chr17_+_82458649 | 0.37 |
ENST00000345415.11
ENST00000412079.6 ENST00000457415.7 ENST00000584411.5 ENST00000577432.5 |
NARF
|
nuclear prelamin A recognition factor |
| chr10_-_14572123 | 0.37 |
ENST00000378465.7
ENST00000452706.6 ENST00000622567.4 ENST00000378458.6 |
FAM107B
|
family with sequence similarity 107 member B |
| chr1_+_81800368 | 0.37 |
ENST00000674489.1
ENST00000674442.1 ENST00000674419.1 ENST00000674407.1 ENST00000674168.1 ENST00000674307.1 ENST00000674209.1 ENST00000370715.5 ENST00000370713.5 ENST00000319517.10 ENST00000627151.2 ENST00000370717.6 |
ADGRL2
|
adhesion G protein-coupled receptor L2 |
| chr19_-_47545261 | 0.36 |
ENST00000595558.1
ENST00000263351.9 |
ZNF541
|
zinc finger protein 541 |
| chr11_-_89491320 | 0.36 |
ENST00000534731.5
ENST00000527626.5 ENST00000525196.5 ENST00000263317.9 |
NOX4
|
NADPH oxidase 4 |
| chr12_+_55931148 | 0.35 |
ENST00000549629.5
ENST00000555218.5 ENST00000331886.10 |
DGKA
|
diacylglycerol kinase alpha |
| chr9_+_113463697 | 0.35 |
ENST00000317613.10
|
RGS3
|
regulator of G protein signaling 3 |
| chr11_-_62707581 | 0.35 |
ENST00000684475.1
ENST00000683296.1 ENST00000684067.1 ENST00000682223.1 |
BSCL2
|
BSCL2 lipid droplet biogenesis associated, seipin |
| chr3_+_137764296 | 0.34 |
ENST00000306087.3
|
SOX14
|
SRY-box transcription factor 14 |
| chr4_-_156970903 | 0.34 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C |
| chr11_-_88175432 | 0.34 |
ENST00000531138.1
ENST00000526372.1 ENST00000243662.11 |
RAB38
|
RAB38, member RAS oncogene family |
| chr7_-_140176970 | 0.34 |
ENST00000397560.7
|
KDM7A
|
lysine demethylase 7A |
| chr19_-_11339573 | 0.33 |
ENST00000222120.8
|
RAB3D
|
RAB3D, member RAS oncogene family |
| chr17_+_82458733 | 0.33 |
ENST00000309794.16
ENST00000577410.2 |
NARF
|
nuclear prelamin A recognition factor |
| chr16_-_4800470 | 0.32 |
ENST00000592019.1
ENST00000586153.1 |
ROGDI
|
rogdi atypical leucine zipper |
| chr8_-_143973520 | 0.32 |
ENST00000356346.7
|
PLEC
|
plectin |
| chr10_-_24952573 | 0.32 |
ENST00000376378.5
ENST00000376376.3 ENST00000320152.11 |
PRTFDC1
|
phosphoribosyl transferase domain containing 1 |
| chr3_+_124094663 | 0.31 |
ENST00000460856.5
ENST00000240874.7 |
KALRN
|
kalirin RhoGEF kinase |
| chr17_+_74432079 | 0.30 |
ENST00000392627.7
ENST00000392628.7 ENST00000582473.2 |
GPRC5C
|
G protein-coupled receptor class C group 5 member C |
| chrX_-_100928903 | 0.30 |
ENST00000372956.3
|
XKRX
|
XK related X-linked |
| chr6_-_32178080 | 0.30 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr14_-_34875348 | 0.27 |
ENST00000360310.6
|
BAZ1A
|
bromodomain adjacent to zinc finger domain 1A |
| chr20_+_18813777 | 0.27 |
ENST00000377428.4
|
SCP2D1
|
SCP2 sterol binding domain containing 1 |
| chr14_-_34874887 | 0.27 |
ENST00000382422.6
|
BAZ1A
|
bromodomain adjacent to zinc finger domain 1A |
| chr2_+_69013337 | 0.27 |
ENST00000463335.2
|
ANTXR1
|
ANTXR cell adhesion molecule 1 |
| chr10_+_7703340 | 0.26 |
ENST00000429820.5
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr17_-_7394800 | 0.26 |
ENST00000574401.5
|
PLSCR3
|
phospholipid scramblase 3 |
| chr22_-_38084093 | 0.26 |
ENST00000681075.1
|
SLC16A8
|
solute carrier family 16 member 8 |
| chr5_-_176537361 | 0.26 |
ENST00000274811.9
|
RNF44
|
ring finger protein 44 |
| chr8_-_33567118 | 0.26 |
ENST00000256257.2
|
RNF122
|
ring finger protein 122 |
| chr16_-_46831134 | 0.26 |
ENST00000394806.6
ENST00000285697.9 |
C16orf87
|
chromosome 16 open reading frame 87 |
| chr7_+_116210501 | 0.25 |
ENST00000455989.1
ENST00000358204.9 |
TES
|
testin LIM domain protein |
| chr17_-_7394514 | 0.25 |
ENST00000571802.1
ENST00000619711.5 ENST00000576201.5 ENST00000573213.1 ENST00000324822.15 |
PLSCR3
|
phospholipid scramblase 3 |
| chr7_+_116672187 | 0.25 |
ENST00000318493.11
ENST00000397752.8 |
MET
|
MET proto-oncogene, receptor tyrosine kinase |
| chr12_+_8914464 | 0.25 |
ENST00000544916.6
|
PHC1
|
polyhomeotic homolog 1 |
| chr18_+_3450036 | 0.24 |
ENST00000546979.5
ENST00000343820.10 ENST00000551402.1 ENST00000577543.5 |
TGIF1
|
TGFB induced factor homeobox 1 |
| chr17_-_7394240 | 0.24 |
ENST00000576362.5
ENST00000571078.5 |
PLSCR3
|
phospholipid scramblase 3 |
| chr14_-_73027077 | 0.24 |
ENST00000553891.5
ENST00000556143.6 |
ZFYVE1
|
zinc finger FYVE-type containing 1 |
| chr7_-_151814636 | 0.23 |
ENST00000652047.1
ENST00000650858.1 |
PRKAG2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chr14_+_105474781 | 0.23 |
ENST00000550577.5
ENST00000538259.2 ENST00000329146.9 |
CRIP2
|
cysteine rich protein 2 |
| chr15_-_55588337 | 0.23 |
ENST00000563719.4
|
PYGO1
|
pygopus family PHD finger 1 |
| chr1_+_182839338 | 0.22 |
ENST00000367549.4
|
DHX9
|
DExH-box helicase 9 |
| chr14_-_73027117 | 0.22 |
ENST00000318876.9
|
ZFYVE1
|
zinc finger FYVE-type containing 1 |
| chr7_-_151814668 | 0.22 |
ENST00000651764.1
|
PRKAG2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chr3_+_52779916 | 0.21 |
ENST00000537050.5
|
ITIH1
|
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr18_+_32190015 | 0.21 |
ENST00000581447.1
|
MEP1B
|
meprin A subunit beta |
| chr18_+_32190033 | 0.21 |
ENST00000269202.11
|
MEP1B
|
meprin A subunit beta |
| chr3_-_126056786 | 0.21 |
ENST00000383598.6
|
SLC41A3
|
solute carrier family 41 member 3 |
| chr10_+_22316445 | 0.21 |
ENST00000448361.5
|
COMMD3
|
COMM domain containing 3 |
| chr12_+_53097656 | 0.21 |
ENST00000301464.4
|
IGFBP6
|
insulin like growth factor binding protein 6 |
| chr15_-_82046119 | 0.20 |
ENST00000558133.1
|
MEX3B
|
mex-3 RNA binding family member B |
| chr10_+_7703300 | 0.20 |
ENST00000358415.9
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr20_+_62817023 | 0.20 |
ENST00000649368.1
|
COL9A3
|
collagen type IX alpha 3 chain |
| chr3_+_138348592 | 0.19 |
ENST00000621127.4
ENST00000494949.5 |
MRAS
|
muscle RAS oncogene homolog |
| chr6_+_108559742 | 0.19 |
ENST00000343882.10
|
FOXO3
|
forkhead box O3 |
| chr20_-_17531366 | 0.19 |
ENST00000377873.8
|
BFSP1
|
beaded filament structural protein 1 |
| chr10_+_22316375 | 0.19 |
ENST00000376836.8
ENST00000456711.5 ENST00000444869.5 ENST00000475460.6 ENST00000602390.5 ENST00000489125.2 |
COMMD3
COMMD3-BMI1
|
COMM domain containing 3 COMMD3-BMI1 readthrough |
| chr7_+_80646305 | 0.19 |
ENST00000426978.5
ENST00000432207.5 |
CD36
|
CD36 molecule |
| chr15_-_82045998 | 0.18 |
ENST00000329713.5
|
MEX3B
|
mex-3 RNA binding family member B |
| chr8_+_119067239 | 0.18 |
ENST00000332843.3
|
COLEC10
|
collectin subfamily member 10 |
| chr13_+_39038347 | 0.18 |
ENST00000379599.6
ENST00000379600.8 |
NHLRC3
|
NHL repeat containing 3 |
| chr5_+_67004618 | 0.18 |
ENST00000261569.11
ENST00000436277.5 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chrX_+_101391202 | 0.18 |
ENST00000471855.1
|
RPL36A
|
ribosomal protein L36a |
| chr4_-_5893075 | 0.18 |
ENST00000324989.12
|
CRMP1
|
collapsin response mediator protein 1 |
| chr13_+_39038292 | 0.18 |
ENST00000470258.5
|
NHLRC3
|
NHL repeat containing 3 |
| chr9_-_120926752 | 0.18 |
ENST00000373887.8
|
TRAF1
|
TNF receptor associated factor 1 |
| chr17_+_77451244 | 0.18 |
ENST00000591088.5
|
SEPTIN9
|
septin 9 |
| chr19_+_10960955 | 0.17 |
ENST00000642628.1
ENST00000643296.1 ENST00000647230.1 |
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr5_-_115180037 | 0.17 |
ENST00000514154.1
ENST00000282369.7 |
TRIM36
|
tripartite motif containing 36 |
| chrX_+_101390976 | 0.17 |
ENST00000392994.7
|
RPL36A
|
ribosomal protein L36a |
| chrX_+_101391000 | 0.17 |
ENST00000553110.8
ENST00000409338.5 ENST00000409170.3 |
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr2_+_233712905 | 0.17 |
ENST00000373414.4
|
UGT1A5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr19_+_10960976 | 0.17 |
ENST00000646510.1
ENST00000429416.8 |
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr10_-_102432565 | 0.17 |
ENST00000369937.5
|
CUEDC2
|
CUE domain containing 2 |
| chr9_-_35650902 | 0.17 |
ENST00000259608.8
ENST00000618781.1 |
SIT1
|
signaling threshold regulating transmembrane adaptor 1 |
| chr15_-_78811415 | 0.16 |
ENST00000388820.5
|
ADAMTS7
|
ADAM metallopeptidase with thrombospondin type 1 motif 7 |
| chr5_+_66828762 | 0.16 |
ENST00000490016.6
ENST00000403666.5 ENST00000450827.5 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr14_-_23155302 | 0.16 |
ENST00000529705.6
|
SLC7A8
|
solute carrier family 7 member 8 |
| chr2_+_178320228 | 0.16 |
ENST00000315022.2
|
OSBPL6
|
oxysterol binding protein like 6 |
| chr11_-_65614195 | 0.16 |
ENST00000309100.8
ENST00000529839.1 ENST00000526293.1 |
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr6_-_11779606 | 0.16 |
ENST00000506810.1
|
ADTRP
|
androgen dependent TFPI regulating protein |
| chr12_+_32107296 | 0.16 |
ENST00000551086.1
|
BICD1
|
BICD cargo adaptor 1 |
| chrX_+_101390824 | 0.16 |
ENST00000427805.6
ENST00000614077.4 |
RPL36A
|
ribosomal protein L36a |
| chr17_+_32486975 | 0.16 |
ENST00000313401.4
|
CDK5R1
|
cyclin dependent kinase 5 regulatory subunit 1 |
| chr1_-_6419903 | 0.16 |
ENST00000377836.8
ENST00000487437.5 ENST00000489730.1 ENST00000377834.8 |
HES2
|
hes family bHLH transcription factor 2 |
| chr12_+_8914525 | 0.16 |
ENST00000543824.5
|
PHC1
|
polyhomeotic homolog 1 |
| chr9_-_72365198 | 0.15 |
ENST00000376962.10
ENST00000376960.8 |
ZFAND5
|
zinc finger AN1-type containing 5 |
| chr5_-_59039454 | 0.15 |
ENST00000358923.10
|
PDE4D
|
phosphodiesterase 4D |
| chr16_-_15643024 | 0.15 |
ENST00000540441.6
|
MARF1
|
meiosis regulator and mRNA stability factor 1 |
| chr8_+_32721823 | 0.15 |
ENST00000539990.3
ENST00000519240.5 |
NRG1
|
neuregulin 1 |
| chr16_-_18801424 | 0.15 |
ENST00000546206.6
ENST00000562819.5 ENST00000304414.12 ENST00000562234.2 ENST00000567078.2 |
ARL6IP1
ENSG00000260342.2
|
ADP ribosylation factor like GTPase 6 interacting protein 1 novel protein |
| chr9_+_100427254 | 0.15 |
ENST00000374885.5
|
MSANTD3
|
Myb/SANT DNA binding domain containing 3 |
| chr18_+_3449817 | 0.15 |
ENST00000407501.6
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chrX_+_136147465 | 0.15 |
ENST00000651929.2
|
FHL1
|
four and a half LIM domains 1 |
| chr6_+_154995258 | 0.15 |
ENST00000682666.1
|
TIAM2
|
TIAM Rac1 associated GEF 2 |
| chr19_+_10960917 | 0.15 |
ENST00000643549.1
ENST00000642726.1 ENST00000646484.1 ENST00000644737.1 |
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr17_-_40937641 | 0.15 |
ENST00000209718.8
|
KRT23
|
keratin 23 |
| chr8_-_42501224 | 0.14 |
ENST00000520262.6
ENST00000517366.1 |
SLC20A2
|
solute carrier family 20 member 2 |
| chr20_-_675793 | 0.14 |
ENST00000488788.2
ENST00000246104.7 |
ENSG00000270299.1
SCRT2
|
novel protein scratch family transcriptional repressor 2 |
| chr8_-_94896660 | 0.14 |
ENST00000520509.5
|
CCNE2
|
cyclin E2 |
| chr7_+_100733576 | 0.14 |
ENST00000613979.5
ENST00000620596.4 |
ZAN
|
zonadhesin |
| chr10_+_11164961 | 0.13 |
ENST00000399850.7
ENST00000417956.6 |
CELF2
|
CUGBP Elav-like family member 2 |
| chr15_-_64703199 | 0.13 |
ENST00000559753.1
ENST00000560258.6 ENST00000559912.2 ENST00000326005.10 |
OAZ2
|
ornithine decarboxylase antizyme 2 |
| chrX_-_136880715 | 0.13 |
ENST00000431446.7
ENST00000320676.11 ENST00000562646.5 |
RBMX
|
RNA binding motif protein X-linked |
| chr17_+_37491464 | 0.13 |
ENST00000613659.1
|
DUSP14
|
dual specificity phosphatase 14 |
| chr9_-_72364504 | 0.13 |
ENST00000237937.7
ENST00000343431.6 ENST00000376956.3 |
ZFAND5
|
zinc finger AN1-type containing 5 |
| chrX_+_126819720 | 0.13 |
ENST00000371125.4
|
PRR32
|
proline rich 32 |
| chr2_-_80304274 | 0.12 |
ENST00000409148.1
ENST00000415098.1 ENST00000452811.1 |
LRRTM1
|
leucine rich repeat transmembrane neuronal 1 |
| chr17_-_40937445 | 0.12 |
ENST00000436344.7
ENST00000485751.1 |
KRT23
|
keratin 23 |
| chr15_-_101252040 | 0.12 |
ENST00000254190.4
|
CHSY1
|
chondroitin sulfate synthase 1 |
| chr14_-_20454741 | 0.12 |
ENST00000206542.9
ENST00000553640.3 |
OSGEP
|
O-sialoglycoprotein endopeptidase |
| chr7_+_80646436 | 0.12 |
ENST00000419819.2
|
CD36
|
CD36 molecule |
| chr1_-_242449478 | 0.12 |
ENST00000427495.5
|
PLD5
|
phospholipase D family member 5 |
| chr17_+_50548041 | 0.12 |
ENST00000634597.1
|
SPATA20
|
spermatogenesis associated 20 |
| chr14_+_75280078 | 0.12 |
ENST00000555347.1
|
FOS
|
Fos proto-oncogene, AP-1 transcription factor subunit |
| chrX_-_154371210 | 0.12 |
ENST00000369856.8
ENST00000422373.6 ENST00000360319.9 |
FLNA
|
filamin A |
| chr17_+_59220446 | 0.12 |
ENST00000284116.9
ENST00000581140.5 ENST00000581276.5 |
GDPD1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr14_-_22982258 | 0.11 |
ENST00000555074.1
ENST00000361265.8 |
ENSG00000259132.1
AJUBA
|
novel protein ajuba LIM protein |
| chr6_-_89217339 | 0.11 |
ENST00000454853.7
|
GABRR1
|
gamma-aminobutyric acid type A receptor subunit rho1 |
| chr14_+_20455185 | 0.11 |
ENST00000555414.5
ENST00000553681.5 |
APEX1
|
apurinic/apyrimidinic endodeoxyribonuclease 1 |
| chr3_+_141387801 | 0.11 |
ENST00000514251.5
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr6_+_95577465 | 0.11 |
ENST00000369293.6
ENST00000683151.1 ENST00000358812.9 |
MANEA
|
mannosidase endo-alpha |
| chr7_+_116672357 | 0.11 |
ENST00000456159.1
|
MET
|
MET proto-oncogene, receptor tyrosine kinase |
| chr15_-_89221558 | 0.11 |
ENST00000268125.10
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr13_+_23570370 | 0.11 |
ENST00000403372.6
ENST00000248484.9 |
TNFRSF19
|
TNF receptor superfamily member 19 |
| chr14_+_20745880 | 0.11 |
ENST00000326842.3
|
EDDM3A
|
epididymal protein 3A |
| chr17_+_59155726 | 0.11 |
ENST00000578777.5
ENST00000577457.1 ENST00000582995.5 ENST00000262293.9 ENST00000614081.1 |
PRR11
|
proline rich 11 |
| chrX_+_136147556 | 0.11 |
ENST00000651089.1
ENST00000420362.5 |
FHL1
|
four and a half LIM domains 1 |
| chr8_+_30095649 | 0.11 |
ENST00000518192.5
|
LEPROTL1
|
leptin receptor overlapping transcript like 1 |
| chr12_+_70366277 | 0.10 |
ENST00000258111.5
|
KCNMB4
|
potassium calcium-activated channel subfamily M regulatory beta subunit 4 |
| chr19_-_48363914 | 0.10 |
ENST00000377431.6
ENST00000293261.8 |
TMEM143
|
transmembrane protein 143 |
| chr9_-_27005659 | 0.10 |
ENST00000380055.6
|
LRRC19
|
leucine rich repeat containing 19 |
| chr1_+_35883189 | 0.10 |
ENST00000674304.1
ENST00000373204.6 ENST00000674426.1 |
AGO1
|
argonaute RISC component 1 |
| chr10_+_113126670 | 0.10 |
ENST00000369389.6
|
TCF7L2
|
transcription factor 7 like 2 |
| chr13_+_75760362 | 0.10 |
ENST00000534657.5
|
LMO7
|
LIM domain 7 |
| chr14_-_22982544 | 0.10 |
ENST00000262713.7
|
AJUBA
|
ajuba LIM protein |
| chr13_+_75760659 | 0.10 |
ENST00000526202.5
ENST00000465261.6 |
LMO7
|
LIM domain 7 |
| chr1_-_42958836 | 0.10 |
ENST00000372500.4
ENST00000674765.1 ENST00000460369.3 ENST00000426263.10 |
SLC2A1
|
solute carrier family 2 member 1 |
| chr1_+_154272589 | 0.09 |
ENST00000457918.6
ENST00000483970.6 ENST00000328703.12 ENST00000435087.1 ENST00000532105.1 |
HAX1
|
HCLS1 associated protein X-1 |
| chr17_+_47522931 | 0.09 |
ENST00000525007.5
ENST00000530173.6 |
NPEPPS
|
aminopeptidase puromycin sensitive |
| chr3_+_141368497 | 0.09 |
ENST00000321464.7
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr8_+_30095400 | 0.09 |
ENST00000321250.13
ENST00000518001.1 ENST00000520682.5 ENST00000442880.6 ENST00000523116.5 |
LEPROTL1
|
leptin receptor overlapping transcript like 1 |
| chr2_+_176129680 | 0.09 |
ENST00000429017.2
ENST00000313173.6 |
HOXD8
|
homeobox D8 |
| chr19_-_48364034 | 0.09 |
ENST00000435956.7
|
TMEM143
|
transmembrane protein 143 |
| chr9_+_136327526 | 0.09 |
ENST00000440944.6
|
GPSM1
|
G protein signaling modulator 1 |
| chr17_+_59565598 | 0.08 |
ENST00000251241.9
ENST00000425628.7 ENST00000584385.5 ENST00000580030.1 |
DHX40
|
DEAH-box helicase 40 |
| chr7_-_19773569 | 0.08 |
ENST00000422233.5
ENST00000433641.5 ENST00000405844.6 |
TMEM196
|
transmembrane protein 196 |
| chr13_+_75760431 | 0.08 |
ENST00000321797.12
|
LMO7
|
LIM domain 7 |
| chr7_+_129188622 | 0.08 |
ENST00000249373.8
|
SMO
|
smoothened, frizzled class receptor |
| chr1_+_109548567 | 0.08 |
ENST00000369851.7
|
GNAI3
|
G protein subunit alpha i3 |
| chr4_+_112231748 | 0.08 |
ENST00000274000.10
ENST00000309703.10 |
AP1AR
|
adaptor related protein complex 1 associated regulatory protein |
| chr7_-_106285898 | 0.08 |
ENST00000424768.2
ENST00000681255.1 |
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr20_-_13990609 | 0.08 |
ENST00000284951.10
ENST00000378072.5 |
SEL1L2
|
SEL1L2 adaptor subunit of ERAD E3 ligase |
| chr20_-_46089905 | 0.07 |
ENST00000372291.3
ENST00000290231.11 |
NCOA5
|
nuclear receptor coactivator 5 |
| chr12_-_48570046 | 0.07 |
ENST00000301046.6
ENST00000549817.1 |
LALBA
|
lactalbumin alpha |
| chr2_-_17800195 | 0.07 |
ENST00000402989.5
ENST00000428868.1 |
SMC6
|
structural maintenance of chromosomes 6 |
| chr13_+_75636311 | 0.07 |
ENST00000377499.9
ENST00000377534.8 |
LMO7
|
LIM domain 7 |
| chr5_-_149551381 | 0.07 |
ENST00000670598.1
ENST00000657001.1 ENST00000515768.6 ENST00000261798.10 ENST00000377843.8 |
CSNK1A1
|
casein kinase 1 alpha 1 |
| chr2_+_26848424 | 0.07 |
ENST00000431402.5
ENST00000614712.4 ENST00000434719.1 |
DPYSL5
|
dihydropyrimidinase like 5 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.2 | 0.7 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.2 | 1.7 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.2 | 0.5 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 | 0.4 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.1 | 0.6 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.3 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.1 | 0.4 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 0.3 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.1 | 0.6 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.3 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.5 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 | 0.2 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.2 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.1 | 0.2 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.1 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.5 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.1 | 0.2 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.0 | 1.5 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.4 | GO:0070543 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.1 | GO:0097698 | telomere maintenance via base-excision repair(GO:0097698) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.2 | GO:0035549 | positive regulation of interferon-beta secretion(GO:0035549) |
| 0.0 | 1.2 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.4 | GO:0060125 | habituation(GO:0046959) negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.1 | GO:1903634 | regulation of calcium-dependent ATPase activity(GO:1903610) negative regulation of calcium-dependent ATPase activity(GO:1903611) regulation of dUTP diphosphatase activity(GO:1903627) positive regulation of dUTP diphosphatase activity(GO:1903629) negative regulation of aminoacyl-tRNA ligase activity(GO:1903631) regulation of leucine-tRNA ligase activity(GO:1903633) negative regulation of leucine-tRNA ligase activity(GO:1903634) |
| 0.0 | 0.1 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.0 | 0.4 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.0 | 0.8 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.5 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.4 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.8 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.6 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 1.4 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.4 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.2 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.2 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.3 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.4 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.6 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.5 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0044334 | canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.0 | 0.2 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.4 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.4 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.7 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.0 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.5 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.4 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.0 | GO:0015744 | succinate transport(GO:0015744) sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) succinate transmembrane transport(GO:0071422) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.2 | 0.5 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 0.5 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 1.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.6 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 0.3 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 1.9 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.1 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.4 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.2 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.4 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.0 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0001939 | female pronucleus(GO:0001939) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.4 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.1 | 0.5 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 1.7 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.4 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.1 | 0.2 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.1 | 0.2 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.1 | 1.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.4 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.4 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0004341 | gluconolactonase activity(GO:0004341) |
| 0.0 | 0.4 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.8 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 1.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.4 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.5 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 2.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.0 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.4 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.1 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |