Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for TEAD4
Z-value: 0.66
Transcription factors associated with TEAD4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TEAD4
|
ENSG00000197905.10 | TEAD4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TEAD4 | hg38_v1_chr12_+_2959296_2959431, hg38_v1_chr12_+_2959870_2959953 | 0.01 | 9.5e-01 | Click! |
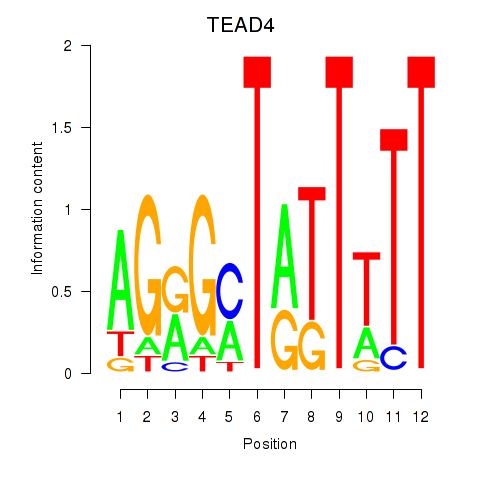
Activity profile of TEAD4 motif
Sorted Z-values of TEAD4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TEAD4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_227813834 | 2.55 |
ENST00000358813.5
ENST00000409189.7 |
CCL20
|
C-C motif chemokine ligand 20 |
| chr4_-_158159657 | 1.24 |
ENST00000590648.5
|
GASK1B
|
golgi associated kinase 1B |
| chr12_-_9760893 | 1.22 |
ENST00000228434.7
ENST00000536709.1 |
CD69
|
CD69 molecule |
| chr4_+_94489030 | 0.93 |
ENST00000510099.5
|
PDLIM5
|
PDZ and LIM domain 5 |
| chr17_+_34255274 | 0.81 |
ENST00000580907.5
ENST00000225831.4 |
CCL2
|
C-C motif chemokine ligand 2 |
| chr2_-_136118142 | 0.79 |
ENST00000241393.4
|
CXCR4
|
C-X-C motif chemokine receptor 4 |
| chr2_-_234497035 | 0.77 |
ENST00000390645.2
ENST00000339728.6 |
ARL4C
|
ADP ribosylation factor like GTPase 4C |
| chr14_+_75605462 | 0.74 |
ENST00000539311.5
|
FLVCR2
|
FLVCR heme transporter 2 |
| chr2_+_200440649 | 0.73 |
ENST00000366118.2
|
SPATS2L
|
spermatogenesis associated serine rich 2 like |
| chr7_-_22194709 | 0.70 |
ENST00000458533.5
|
RAPGEF5
|
Rap guanine nucleotide exchange factor 5 |
| chr7_-_36985060 | 0.69 |
ENST00000396040.6
|
ELMO1
|
engulfment and cell motility 1 |
| chr3_+_101827982 | 0.69 |
ENST00000461724.5
ENST00000483180.5 ENST00000394054.6 |
NFKBIZ
|
NFKB inhibitor zeta |
| chr11_-_128587551 | 0.68 |
ENST00000392668.8
|
ETS1
|
ETS proto-oncogene 1, transcription factor |
| chr4_+_41612702 | 0.67 |
ENST00000509277.5
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr4_+_41612892 | 0.60 |
ENST00000509454.5
ENST00000396595.7 ENST00000381753.8 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr3_-_185821092 | 0.59 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2 |
| chr2_+_151357583 | 0.55 |
ENST00000243347.5
|
TNFAIP6
|
TNF alpha induced protein 6 |
| chr5_-_32312913 | 0.55 |
ENST00000280285.9
ENST00000382142.8 ENST00000264934.5 |
MTMR12
|
myotubularin related protein 12 |
| chr10_-_49762276 | 0.50 |
ENST00000374103.9
|
OGDHL
|
oxoglutarate dehydrogenase L |
| chr10_-_49762335 | 0.48 |
ENST00000419399.4
ENST00000432695.2 |
OGDHL
|
oxoglutarate dehydrogenase L |
| chr11_-_118925916 | 0.47 |
ENST00000683865.1
|
BCL9L
|
BCL9 like |
| chr12_-_24903014 | 0.46 |
ENST00000539282.5
|
BCAT1
|
branched chain amino acid transaminase 1 |
| chr6_-_159726871 | 0.45 |
ENST00000535561.5
|
SOD2
|
superoxide dismutase 2 |
| chr7_+_77538027 | 0.44 |
ENST00000433369.6
ENST00000415482.6 |
PTPN12
|
protein tyrosine phosphatase non-receptor type 12 |
| chr9_+_72577369 | 0.43 |
ENST00000651183.1
|
TMC1
|
transmembrane channel like 1 |
| chr1_+_65992389 | 0.43 |
ENST00000423207.6
|
PDE4B
|
phosphodiesterase 4B |
| chr10_+_84452208 | 0.41 |
ENST00000480006.1
|
CCSER2
|
coiled-coil serine rich protein 2 |
| chr8_+_22367526 | 0.40 |
ENST00000289952.9
ENST00000524285.1 |
SLC39A14
|
solute carrier family 39 member 14 |
| chr9_+_128566741 | 0.39 |
ENST00000630866.1
|
SPTAN1
|
spectrin alpha, non-erythrocytic 1 |
| chr15_-_68820861 | 0.38 |
ENST00000560303.1
ENST00000465139.6 |
ANP32A
|
acidic nuclear phosphoprotein 32 family member A |
| chr3_+_112086335 | 0.36 |
ENST00000431717.6
ENST00000480282.5 |
C3orf52
|
chromosome 3 open reading frame 52 |
| chr4_-_88231322 | 0.36 |
ENST00000515655.5
|
ABCG2
|
ATP binding cassette subfamily G member 2 (Junior blood group) |
| chr14_+_30577752 | 0.35 |
ENST00000547532.5
ENST00000555429.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr8_+_106447918 | 0.35 |
ENST00000442977.6
|
OXR1
|
oxidation resistance 1 |
| chr7_-_82443715 | 0.35 |
ENST00000356253.9
ENST00000423588.1 |
CACNA2D1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr9_+_113594118 | 0.34 |
ENST00000620489.1
|
RGS3
|
regulator of G protein signaling 3 |
| chrX_+_21374288 | 0.34 |
ENST00000642359.1
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr2_-_187554473 | 0.34 |
ENST00000453013.5
ENST00000417013.5 |
TFPI
|
tissue factor pathway inhibitor |
| chr9_-_127847117 | 0.32 |
ENST00000480266.5
|
ENG
|
endoglin |
| chr4_+_185395979 | 0.30 |
ENST00000507753.1
|
ANKRD37
|
ankyrin repeat domain 37 |
| chr11_+_102347312 | 0.29 |
ENST00000621637.1
ENST00000613397.4 |
BIRC2
|
baculoviral IAP repeat containing 2 |
| chr17_-_18682262 | 0.29 |
ENST00000454745.2
ENST00000395675.7 |
FOXO3B
|
forkhead box O3B |
| chr6_-_117573571 | 0.29 |
ENST00000467125.1
|
ENSG00000282218.1
|
novel protein, GOPC-ROS1 readthrough |
| chr6_-_41071825 | 0.29 |
ENST00000468811.5
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr17_+_42458844 | 0.29 |
ENST00000393829.6
ENST00000537728.5 ENST00000343619.9 ENST00000264649.10 ENST00000585525.5 ENST00000544137.5 ENST00000589727.5 ENST00000587824.5 |
ATP6V0A1
|
ATPase H+ transporting V0 subunit a1 |
| chr5_+_139308095 | 0.29 |
ENST00000515833.2
|
MATR3
|
matrin 3 |
| chr1_+_154257071 | 0.28 |
ENST00000428595.1
|
UBAP2L
|
ubiquitin associated protein 2 like |
| chr8_-_23854796 | 0.28 |
ENST00000290271.7
|
STC1
|
stanniocalcin 1 |
| chr5_-_95081482 | 0.27 |
ENST00000312216.12
ENST00000512425.5 ENST00000505208.5 ENST00000429576.6 ENST00000508509.5 ENST00000510732.5 |
MCTP1
|
multiple C2 and transmembrane domain containing 1 |
| chr22_-_50085414 | 0.27 |
ENST00000311597.10
|
MLC1
|
modulator of VRAC current 1 |
| chr2_+_237858874 | 0.26 |
ENST00000404910.6
|
RAMP1
|
receptor activity modifying protein 1 |
| chr2_+_108288869 | 0.26 |
ENST00000251481.11
ENST00000438339.5 ENST00000409880.5 ENST00000437390.6 |
SULT1C2
|
sulfotransferase family 1C member 2 |
| chr10_-_73651013 | 0.26 |
ENST00000372873.8
|
SYNPO2L
|
synaptopodin 2 like |
| chr22_-_50085331 | 0.26 |
ENST00000395876.6
|
MLC1
|
modulator of VRAC current 1 |
| chr1_-_154956086 | 0.26 |
ENST00000368463.8
ENST00000368460.7 ENST00000368465.5 |
PBXIP1
|
PBX homeobox interacting protein 1 |
| chr11_-_118264282 | 0.26 |
ENST00000278937.7
|
MPZL2
|
myelin protein zero like 2 |
| chr3_-_18424533 | 0.25 |
ENST00000417717.6
|
SATB1
|
SATB homeobox 1 |
| chr3_-_71132099 | 0.24 |
ENST00000650188.1
ENST00000648121.1 ENST00000648794.1 ENST00000649592.1 |
FOXP1
|
forkhead box P1 |
| chr6_-_42722590 | 0.24 |
ENST00000230381.7
|
PRPH2
|
peripherin 2 |
| chr17_-_68955332 | 0.24 |
ENST00000269080.6
ENST00000615593.4 ENST00000586539.6 ENST00000430352.6 |
ABCA8
|
ATP binding cassette subfamily A member 8 |
| chr3_-_197260369 | 0.24 |
ENST00000658155.1
ENST00000453607.5 |
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr1_+_241532370 | 0.23 |
ENST00000366559.9
ENST00000366557.8 |
KMO
|
kynurenine 3-monooxygenase |
| chrX_+_21374608 | 0.23 |
ENST00000644295.1
ENST00000645074.1 ENST00000645791.1 ENST00000643220.1 |
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr4_-_185810894 | 0.22 |
ENST00000448662.6
ENST00000439049.5 ENST00000420158.5 ENST00000319471.13 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr6_-_52840843 | 0.22 |
ENST00000370989.6
|
GSTA5
|
glutathione S-transferase alpha 5 |
| chr6_-_46325641 | 0.21 |
ENST00000330430.10
ENST00000405162.2 |
RCAN2
|
regulator of calcineurin 2 |
| chr1_+_89633086 | 0.21 |
ENST00000370454.9
|
LRRC8C
|
leucine rich repeat containing 8 VRAC subunit C |
| chr17_-_40937641 | 0.21 |
ENST00000209718.8
|
KRT23
|
keratin 23 |
| chr2_-_231125032 | 0.21 |
ENST00000258400.4
|
HTR2B
|
5-hydroxytryptamine receptor 2B |
| chr4_+_95091462 | 0.21 |
ENST00000264568.8
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr20_+_44531817 | 0.20 |
ENST00000372889.5
ENST00000372887.5 |
PKIG
|
cAMP-dependent protein kinase inhibitor gamma |
| chr8_+_66493514 | 0.20 |
ENST00000521495.5
|
VXN
|
vexin |
| chr3_-_71130963 | 0.20 |
ENST00000649695.2
|
FOXP1
|
forkhead box P1 |
| chr4_-_109801978 | 0.19 |
ENST00000510800.1
ENST00000512148.5 ENST00000394634.7 ENST00000394635.8 ENST00000645635.1 |
CFI
ENSG00000285330.1
|
complement factor I novel protein |
| chr13_-_37105664 | 0.19 |
ENST00000379800.4
|
CSNK1A1L
|
casein kinase 1 alpha 1 like |
| chr6_-_132659178 | 0.19 |
ENST00000275216.3
|
TAAR1
|
trace amine associated receptor 1 |
| chr18_+_6834473 | 0.19 |
ENST00000581099.5
ENST00000419673.6 ENST00000531294.5 |
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr7_+_77538059 | 0.19 |
ENST00000435495.6
|
PTPN12
|
protein tyrosine phosphatase non-receptor type 12 |
| chr4_-_144140712 | 0.19 |
ENST00000641688.3
|
GYPA
|
glycophorin A (MNS blood group) |
| chr6_+_112236806 | 0.19 |
ENST00000588837.5
ENST00000590293.5 ENST00000585450.5 ENST00000629766.2 ENST00000590804.5 ENST00000590584.4 ENST00000628122.2 ENST00000627025.1 ENST00000590673.5 ENST00000585611.5 ENST00000587816.2 |
LAMA4-AS1
ENSG00000281613.2
|
LAMA4 antisense RNA 1 novel ret finger protein-like 4B |
| chr15_-_70702273 | 0.18 |
ENST00000558758.5
ENST00000379983.6 ENST00000560441.5 |
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr12_+_18262730 | 0.18 |
ENST00000675017.1
|
PIK3C2G
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
| chr4_-_144140725 | 0.18 |
ENST00000360771.8
|
GYPA
|
glycophorin A (MNS blood group) |
| chr2_-_196068812 | 0.18 |
ENST00000410072.5
ENST00000312428.11 |
DNAH7
|
dynein axonemal heavy chain 7 |
| chr3_-_71130557 | 0.18 |
ENST00000497355.7
|
FOXP1
|
forkhead box P1 |
| chr12_-_57826295 | 0.18 |
ENST00000549039.5
|
CTDSP2
|
CTD small phosphatase 2 |
| chr2_+_190927649 | 0.17 |
ENST00000409428.5
ENST00000409215.5 |
GLS
|
glutaminase |
| chr15_+_59438149 | 0.17 |
ENST00000288228.10
ENST00000559628.5 ENST00000557914.5 ENST00000560474.5 |
FAM81A
|
family with sequence similarity 81 member A |
| chr7_+_54542393 | 0.17 |
ENST00000404951.5
|
VSTM2A
|
V-set and transmembrane domain containing 2A |
| chr6_+_159726998 | 0.17 |
ENST00000614346.4
|
WTAP
|
WT1 associated protein |
| chr12_-_91178520 | 0.17 |
ENST00000425043.5
ENST00000420120.6 ENST00000441303.6 ENST00000456569.2 |
DCN
|
decorin |
| chr17_+_81395469 | 0.17 |
ENST00000584436.7
|
BAHCC1
|
BAH domain and coiled-coil containing 1 |
| chr16_-_20669855 | 0.16 |
ENST00000524149.5
|
ACSM1
|
acyl-CoA synthetase medium chain family member 1 |
| chr3_-_71130892 | 0.16 |
ENST00000491238.7
ENST00000674446.1 |
FOXP1
|
forkhead box P1 |
| chr14_+_21797272 | 0.16 |
ENST00000390430.2
|
TRAV8-1
|
T cell receptor alpha variable 8-1 |
| chr17_+_81395449 | 0.16 |
ENST00000675386.2
|
BAHCC1
|
BAH domain and coiled-coil containing 1 |
| chr18_+_58195390 | 0.16 |
ENST00000456173.6
ENST00000676226.1 ENST00000675865.1 |
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr1_+_160796157 | 0.16 |
ENST00000263285.11
ENST00000368039.2 |
LY9
|
lymphocyte antigen 9 |
| chr18_-_3219961 | 0.15 |
ENST00000356443.9
|
MYOM1
|
myomesin 1 |
| chrX_+_37780049 | 0.15 |
ENST00000378588.5
|
CYBB
|
cytochrome b-245 beta chain |
| chr8_-_69833338 | 0.15 |
ENST00000524945.5
|
SLCO5A1
|
solute carrier organic anion transporter family member 5A1 |
| chr7_+_54542362 | 0.15 |
ENST00000402613.4
|
VSTM2A
|
V-set and transmembrane domain containing 2A |
| chr7_+_54542300 | 0.15 |
ENST00000302287.7
ENST00000407838.7 |
VSTM2A
|
V-set and transmembrane domain containing 2A |
| chr17_-_40937445 | 0.14 |
ENST00000436344.7
ENST00000485751.1 |
KRT23
|
keratin 23 |
| chr6_-_166168700 | 0.14 |
ENST00000366871.7
|
TBXT
|
T-box transcription factor T |
| chr16_-_29862890 | 0.14 |
ENST00000563415.1
|
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chr18_-_55422306 | 0.14 |
ENST00000566777.5
ENST00000626584.2 |
TCF4
|
transcription factor 4 |
| chr3_+_14675128 | 0.14 |
ENST00000435614.5
ENST00000253697.8 ENST00000412910.1 |
C3orf20
|
chromosome 3 open reading frame 20 |
| chrX_+_21374434 | 0.14 |
ENST00000279451.9
ENST00000645245.1 |
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr6_-_166168612 | 0.14 |
ENST00000296946.6
|
TBXT
|
T-box transcription factor T |
| chr2_-_157327699 | 0.13 |
ENST00000397283.6
|
ERMN
|
ermin |
| chr1_-_225999312 | 0.13 |
ENST00000272091.8
|
SDE2
|
SDE2 telomere maintenance homolog |
| chr2_-_60553558 | 0.13 |
ENST00000642439.1
ENST00000356842.9 |
BCL11A
|
BAF chromatin remodeling complex subunit BCL11A |
| chr2_-_160200310 | 0.13 |
ENST00000620391.4
|
ITGB6
|
integrin subunit beta 6 |
| chr15_+_99566066 | 0.12 |
ENST00000557942.5
|
MEF2A
|
myocyte enhancer factor 2A |
| chr20_+_35226676 | 0.12 |
ENST00000246186.8
|
MMP24
|
matrix metallopeptidase 24 |
| chr7_-_47948448 | 0.12 |
ENST00000289672.7
|
PKD1L1
|
polycystin 1 like 1, transient receptor potential channel interacting |
| chr3_-_112638097 | 0.12 |
ENST00000461431.1
|
CCDC80
|
coiled-coil domain containing 80 |
| chr12_-_122526929 | 0.12 |
ENST00000331738.12
ENST00000528279.1 ENST00000344591.8 ENST00000526560.6 |
RSRC2
|
arginine and serine rich coiled-coil 2 |
| chr11_-_55936400 | 0.12 |
ENST00000301532.3
|
OR5I1
|
olfactory receptor family 5 subfamily I member 1 |
| chr12_-_21604840 | 0.12 |
ENST00000261195.3
|
GYS2
|
glycogen synthase 2 |
| chr3_-_71360753 | 0.12 |
ENST00000648783.1
|
FOXP1
|
forkhead box P1 |
| chr20_+_5750437 | 0.12 |
ENST00000445603.1
ENST00000442185.1 |
SHLD1
|
shieldin complex subunit 1 |
| chr5_+_173889337 | 0.12 |
ENST00000520867.5
ENST00000334035.9 |
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr4_-_39032343 | 0.12 |
ENST00000381938.4
|
TMEM156
|
transmembrane protein 156 |
| chr9_+_122510802 | 0.12 |
ENST00000335302.5
|
OR1J2
|
olfactory receptor family 1 subfamily J member 2 |
| chr19_+_35139440 | 0.12 |
ENST00000455515.6
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr12_+_32107296 | 0.12 |
ENST00000551086.1
|
BICD1
|
BICD cargo adaptor 1 |
| chr15_+_99565950 | 0.12 |
ENST00000557785.5
ENST00000558049.5 ENST00000449277.6 |
MEF2A
|
myocyte enhancer factor 2A |
| chr15_+_99565921 | 0.12 |
ENST00000558812.5
ENST00000338042.10 |
MEF2A
|
myocyte enhancer factor 2A |
| chr21_+_46286623 | 0.12 |
ENST00000397691.1
|
YBEY
|
ybeY metalloendoribonuclease |
| chr3_+_138348823 | 0.12 |
ENST00000475711.5
ENST00000464896.5 |
MRAS
|
muscle RAS oncogene homolog |
| chr6_+_108848387 | 0.11 |
ENST00000368972.7
ENST00000392644.9 |
ARMC2
|
armadillo repeat containing 2 |
| chr19_+_35139724 | 0.11 |
ENST00000588715.5
ENST00000588607.5 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr2_-_187554351 | 0.11 |
ENST00000437725.5
ENST00000409676.5 ENST00000233156.9 ENST00000339091.8 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor |
| chr1_-_11848345 | 0.11 |
ENST00000376476.1
|
NPPA
|
natriuretic peptide A |
| chr2_-_160200251 | 0.11 |
ENST00000428609.6
ENST00000409967.6 ENST00000283249.7 |
ITGB6
|
integrin subunit beta 6 |
| chr1_+_241532121 | 0.10 |
ENST00000366558.7
|
KMO
|
kynurenine 3-monooxygenase |
| chr12_-_54258275 | 0.10 |
ENST00000552562.1
|
CBX5
|
chromobox 5 |
| chr18_+_58341038 | 0.10 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr6_+_31655888 | 0.10 |
ENST00000375916.4
|
APOM
|
apolipoprotein M |
| chr6_+_31670379 | 0.10 |
ENST00000409525.1
|
LY6G5B
|
lymphocyte antigen 6 family member G5B |
| chr3_+_130560334 | 0.09 |
ENST00000358511.10
|
COL6A6
|
collagen type VI alpha 6 chain |
| chr7_-_108240049 | 0.09 |
ENST00000379022.8
|
NRCAM
|
neuronal cell adhesion molecule |
| chr5_-_180072086 | 0.09 |
ENST00000261947.4
|
RNF130
|
ring finger protein 130 |
| chr11_+_62242232 | 0.09 |
ENST00000244926.4
|
SCGB1D2
|
secretoglobin family 1D member 2 |
| chrX_-_54998530 | 0.09 |
ENST00000545676.5
|
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr1_+_15684284 | 0.09 |
ENST00000375799.8
ENST00000375793.2 |
PLEKHM2
|
pleckstrin homology and RUN domain containing M2 |
| chr19_+_10013468 | 0.09 |
ENST00000591589.3
|
RDH8
|
retinol dehydrogenase 8 |
| chr5_+_54455661 | 0.09 |
ENST00000302005.3
|
HSPB3
|
heat shock protein family B (small) member 3 |
| chr21_-_46285608 | 0.09 |
ENST00000291688.6
|
MCM3AP
|
minichromosome maintenance complex component 3 associated protein |
| chr17_+_17782108 | 0.08 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr6_-_87702221 | 0.08 |
ENST00000257787.6
|
AKIRIN2
|
akirin 2 |
| chrX_+_21374357 | 0.08 |
ENST00000643841.1
ENST00000379510.5 ENST00000425654.7 ENST00000644798.1 ENST00000543067.6 |
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chrX_+_80420466 | 0.08 |
ENST00000308293.5
|
TENT5D
|
terminal nucleotidyltransferase 5D |
| chr19_+_35138993 | 0.08 |
ENST00000612146.4
ENST00000589209.5 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr12_-_91146195 | 0.07 |
ENST00000548218.1
|
DCN
|
decorin |
| chr3_-_11643871 | 0.07 |
ENST00000430365.7
|
VGLL4
|
vestigial like family member 4 |
| chr11_+_46936710 | 0.07 |
ENST00000378618.6
ENST00000278460.12 ENST00000395460.6 ENST00000378615.7 |
C11orf49
|
chromosome 11 open reading frame 49 |
| chr11_+_6845683 | 0.07 |
ENST00000299454.5
|
OR10A5
|
olfactory receptor family 10 subfamily A member 5 |
| chr3_-_177197248 | 0.07 |
ENST00000427349.5
ENST00000352800.10 |
TBL1XR1
|
TBL1X receptor 1 |
| chr14_-_106803221 | 0.07 |
ENST00000390636.2
|
IGHV3-73
|
immunoglobulin heavy variable 3-73 |
| chr8_-_27258386 | 0.07 |
ENST00000350889.8
ENST00000519997.5 ENST00000519614.5 ENST00000522908.1 ENST00000265770.11 |
STMN4
|
stathmin 4 |
| chr13_-_44437214 | 0.07 |
ENST00000622051.1
|
TSC22D1
|
TSC22 domain family member 1 |
| chr10_+_15032193 | 0.06 |
ENST00000428897.5
ENST00000413672.5 |
OLAH
|
oleoyl-ACP hydrolase |
| chr9_+_68356899 | 0.06 |
ENST00000396392.5
|
PGM5
|
phosphoglucomutase 5 |
| chr6_-_152168349 | 0.06 |
ENST00000539504.5
|
SYNE1
|
spectrin repeat containing nuclear envelope protein 1 |
| chr20_+_5750381 | 0.06 |
ENST00000378979.8
ENST00000303142.11 |
SHLD1
|
shieldin complex subunit 1 |
| chr1_+_43269974 | 0.06 |
ENST00000439858.6
|
TMEM125
|
transmembrane protein 125 |
| chr4_+_122239965 | 0.06 |
ENST00000446180.5
|
KIAA1109
|
KIAA1109 |
| chr2_-_160200289 | 0.06 |
ENST00000409872.1
|
ITGB6
|
integrin subunit beta 6 |
| chr6_-_152168291 | 0.06 |
ENST00000354674.5
|
SYNE1
|
spectrin repeat containing nuclear envelope protein 1 |
| chr20_-_13990609 | 0.06 |
ENST00000284951.10
ENST00000378072.5 |
SEL1L2
|
SEL1L2 adaptor subunit of ERAD E3 ligase |
| chr11_+_126327863 | 0.06 |
ENST00000648516.1
|
DCPS
|
decapping enzyme, scavenger |
| chr1_-_212847649 | 0.06 |
ENST00000332912.3
|
SPATA45
|
spermatogenesis associated 45 |
| chr21_-_38498415 | 0.06 |
ENST00000398905.5
ENST00000398907.5 ENST00000453032.6 ENST00000288319.12 |
ERG
|
ETS transcription factor ERG |
| chr1_+_43270007 | 0.05 |
ENST00000432792.6
|
TMEM125
|
transmembrane protein 125 |
| chr3_-_15798184 | 0.05 |
ENST00000624145.3
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr12_-_39619782 | 0.05 |
ENST00000308666.4
|
ABCD2
|
ATP binding cassette subfamily D member 2 |
| chr10_+_27532521 | 0.05 |
ENST00000683924.1
|
RAB18
|
RAB18, member RAS oncogene family |
| chr6_-_76072654 | 0.05 |
ENST00000369950.8
ENST00000611179.4 ENST00000369963.5 |
IMPG1
|
interphotoreceptor matrix proteoglycan 1 |
| chr12_+_10307950 | 0.05 |
ENST00000543420.5
ENST00000543777.5 |
KLRD1
|
killer cell lectin like receptor D1 |
| chr17_+_40062956 | 0.05 |
ENST00000450525.7
|
THRA
|
thyroid hormone receptor alpha |
| chr7_-_93148345 | 0.05 |
ENST00000437805.5
ENST00000446959.5 ENST00000439952.5 ENST00000414791.5 ENST00000446033.1 ENST00000411955.5 ENST00000318238.9 |
SAMD9L
|
sterile alpha motif domain containing 9 like |
| chr7_+_148133684 | 0.05 |
ENST00000628930.2
|
CNTNAP2
|
contactin associated protein 2 |
| chr6_+_31670167 | 0.05 |
ENST00000375864.4
|
LY6G5B
|
lymphocyte antigen 6 family member G5B |
| chr10_+_62049211 | 0.05 |
ENST00000309334.5
|
ARID5B
|
AT-rich interaction domain 5B |
| chr1_+_43530847 | 0.05 |
ENST00000617451.4
ENST00000359947.9 ENST00000438120.5 |
PTPRF
|
protein tyrosine phosphatase receptor type F |
| chr9_+_68356603 | 0.05 |
ENST00000396396.6
|
PGM5
|
phosphoglucomutase 5 |
| chr1_+_40738834 | 0.05 |
ENST00000525290.5
ENST00000530965.5 ENST00000416859.6 ENST00000308733.9 |
NFYC
|
nuclear transcription factor Y subunit gamma |
| chr9_-_71121596 | 0.05 |
ENST00000377110.9
ENST00000377111.8 ENST00000677713.2 |
TRPM3
|
transient receptor potential cation channel subfamily M member 3 |
| chr12_+_80099535 | 0.05 |
ENST00000646859.1
ENST00000547103.7 |
OTOGL
|
otogelin like |
| chr10_+_24239181 | 0.05 |
ENST00000438429.5
|
KIAA1217
|
KIAA1217 |
| chr8_-_27258414 | 0.05 |
ENST00000523048.5
|
STMN4
|
stathmin 4 |
| chr3_+_186570663 | 0.04 |
ENST00000265028.8
|
DNAJB11
|
DnaJ heat shock protein family (Hsp40) member B11 |
| chr11_-_84273188 | 0.04 |
ENST00000330014.11
ENST00000418306.6 ENST00000531015.5 |
DLG2
|
discs large MAGUK scaffold protein 2 |
| chr11_+_9575496 | 0.04 |
ENST00000681684.1
|
WEE1
|
WEE1 G2 checkpoint kinase |
| chr8_+_42271289 | 0.04 |
ENST00000520810.6
ENST00000520835.7 |
IKBKB
|
inhibitor of nuclear factor kappa B kinase subunit beta |
| chr8_+_66493556 | 0.04 |
ENST00000305454.8
ENST00000522977.5 ENST00000480005.1 |
VXN
|
vexin |
| chr16_+_32995048 | 0.04 |
ENST00000425181.3
|
IGHV3OR16-10
|
immunoglobulin heavy variable 3/OR16-10 (non-functional) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.3 | 0.8 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.1 | 0.5 | GO:0070352 | positive regulation of white fat cell proliferation(GO:0070352) |
| 0.1 | 0.3 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.1 | 0.3 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.1 | 0.5 | GO:0009098 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 | 1.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.5 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 0.8 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 0.3 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.1 | 0.3 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.7 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.1 | 0.2 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.1 | 0.9 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.4 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 0.3 | GO:0010731 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.0 | 0.4 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.4 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.5 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.2 | GO:0010513 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.4 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.5 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 1.3 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.3 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.2 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.0 | 0.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.4 | GO:0015684 | ferrous iron transport(GO:0015684) ferrous iron transmembrane transport(GO:1903874) |
| 0.0 | 0.3 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.3 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.1 | GO:0009726 | detection of endogenous stimulus(GO:0009726) |
| 0.0 | 0.1 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.0 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 1.0 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.2 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:2000173 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.2 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.3 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.8 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.7 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.0 | GO:1903347 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 1.2 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.1 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.1 | 1.0 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.3 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.2 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.6 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.3 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.2 | 0.8 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.2 | 1.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 1.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.5 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.3 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 0.8 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.3 | GO:0031716 | calcitonin receptor activity(GO:0004948) calcitonin receptor binding(GO:0031716) |
| 0.0 | 0.4 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.7 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.5 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.6 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.3 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.4 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.9 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.3 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.8 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 1.0 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0016295 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.3 | GO:0019871 | potassium channel inhibitor activity(GO:0019870) sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 3.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.7 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |