Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
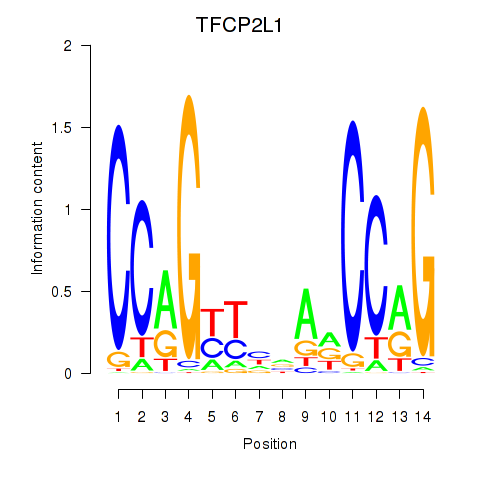
Results for TFCP2L1
Z-value: 0.82
Transcription factors associated with TFCP2L1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFCP2L1
|
ENSG00000115112.8 | TFCP2L1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TFCP2L1 | hg38_v1_chr2_-_121285194_121285209 | 0.25 | 2.3e-01 | Click! |
Activity profile of TFCP2L1 motif
Sorted Z-values of TFCP2L1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TFCP2L1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_19712823 | 2.69 |
ENST00000396085.6
ENST00000349880.9 |
NAV2
|
neuron navigator 2 |
| chr6_-_31582415 | 2.07 |
ENST00000429299.3
ENST00000446745.2 |
LTB
|
lymphotoxin beta |
| chr8_+_53851786 | 1.74 |
ENST00000297313.8
ENST00000344277.10 |
RGS20
|
regulator of G protein signaling 20 |
| chr2_-_10448318 | 1.73 |
ENST00000234111.9
|
ODC1
|
ornithine decarboxylase 1 |
| chr15_-_35085295 | 1.62 |
ENST00000528386.4
|
NANOGP8
|
Nanog homeobox retrogene P8 |
| chr16_-_65121930 | 1.41 |
ENST00000566827.5
ENST00000394156.7 ENST00000268603.9 ENST00000562998.1 |
CDH11
|
cadherin 11 |
| chr1_+_205256189 | 1.40 |
ENST00000329800.7
|
TMCC2
|
transmembrane and coiled-coil domain family 2 |
| chr14_+_96204679 | 1.33 |
ENST00000542454.2
ENST00000539359.1 ENST00000554311.2 ENST00000553811.1 |
BDKRB2
ENSG00000258691.1
|
bradykinin receptor B2 novel protein |
| chr19_-_4338786 | 1.28 |
ENST00000601482.1
ENST00000600324.5 ENST00000594605.6 |
STAP2
|
signal transducing adaptor family member 2 |
| chr7_+_151341764 | 1.26 |
ENST00000413040.7
ENST00000470229.6 ENST00000568733.6 |
NUB1
|
negative regulator of ubiquitin like proteins 1 |
| chr1_+_205257180 | 1.24 |
ENST00000330675.12
|
TMCC2
|
transmembrane and coiled-coil domain family 2 |
| chr21_-_15064934 | 1.14 |
ENST00000400199.5
ENST00000400202.5 ENST00000318948.7 |
NRIP1
|
nuclear receptor interacting protein 1 |
| chr7_+_66205325 | 1.12 |
ENST00000304842.6
ENST00000649664.1 |
TPST1
|
tyrosylprotein sulfotransferase 1 |
| chr16_-_11587162 | 1.09 |
ENST00000570904.5
ENST00000574701.5 |
LITAF
|
lipopolysaccharide induced TNF factor |
| chr22_-_37427433 | 1.06 |
ENST00000452946.1
ENST00000402918.7 |
ELFN2
ELFN2
|
extracellular leucine rich repeat and fibronectin type III domain containing 2 extracellular leucine rich repeat and fibronectin type III domain containing 2 |
| chr12_-_48004496 | 1.00 |
ENST00000337299.7
|
COL2A1
|
collagen type II alpha 1 chain |
| chr1_-_111200633 | 0.97 |
ENST00000357640.9
|
DENND2D
|
DENN domain containing 2D |
| chr3_+_173584433 | 0.96 |
ENST00000361589.8
|
NLGN1
|
neuroligin 1 |
| chr6_+_31587185 | 0.96 |
ENST00000376092.7
ENST00000376086.7 ENST00000303757.12 ENST00000376093.6 |
LST1
|
leukocyte specific transcript 1 |
| chr14_+_32077068 | 0.94 |
ENST00000396582.6
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr10_-_73625951 | 0.91 |
ENST00000433394.1
ENST00000422491.7 |
USP54
|
ubiquitin specific peptidase 54 |
| chr12_-_121274510 | 0.80 |
ENST00000392474.6
|
CAMKK2
|
calcium/calmodulin dependent protein kinase kinase 2 |
| chr9_+_88991440 | 0.77 |
ENST00000358157.3
|
S1PR3
|
sphingosine-1-phosphate receptor 3 |
| chr20_+_56629296 | 0.73 |
ENST00000201031.3
|
TFAP2C
|
transcription factor AP-2 gamma |
| chr10_-_119536533 | 0.72 |
ENST00000392865.5
|
RGS10
|
regulator of G protein signaling 10 |
| chr16_+_84294853 | 0.72 |
ENST00000219454.10
|
WFDC1
|
WAP four-disulfide core domain 1 |
| chr3_-_49929800 | 0.71 |
ENST00000455683.6
|
MON1A
|
MON1 homolog A, secretory trafficking associated |
| chr16_+_84294823 | 0.70 |
ENST00000568638.1
|
WFDC1
|
WAP four-disulfide core domain 1 |
| chr2_+_46941199 | 0.70 |
ENST00000319190.11
ENST00000394850.6 |
TTC7A
|
tetratricopeptide repeat domain 7A |
| chr19_-_12140315 | 0.64 |
ENST00000418866.1
ENST00000600335.5 ENST00000334213.10 |
ZNF20
|
zinc finger protein 20 |
| chr14_+_32077280 | 0.64 |
ENST00000432921.5
ENST00000345122.8 ENST00000433497.5 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr1_-_201115372 | 0.62 |
ENST00000458416.2
|
ASCL5
|
achaete-scute family bHLH transcription factor 5 |
| chr11_+_96389985 | 0.58 |
ENST00000332349.5
|
JRKL
|
JRK like |
| chr3_-_50340933 | 0.58 |
ENST00000616212.4
|
RASSF1
|
Ras association domain family member 1 |
| chr11_+_62881686 | 0.58 |
ENST00000536981.6
ENST00000539891.6 |
SLC3A2
|
solute carrier family 3 member 2 |
| chr3_-_50340804 | 0.56 |
ENST00000359365.9
ENST00000357043.6 |
RASSF1
|
Ras association domain family member 1 |
| chr20_-_44651683 | 0.56 |
ENST00000537820.1
ENST00000372874.9 |
ADA
|
adenosine deaminase |
| chr10_+_122163426 | 0.56 |
ENST00000360561.7
|
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chr1_-_161550591 | 0.56 |
ENST00000367967.7
ENST00000436743.6 ENST00000442336.1 |
FCGR3A
|
Fc fragment of IgG receptor IIIa |
| chr10_+_122163590 | 0.55 |
ENST00000368999.5
|
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chr7_-_113086150 | 0.54 |
ENST00000449591.2
|
GPR85
|
G protein-coupled receptor 85 |
| chr10_+_122163672 | 0.53 |
ENST00000369004.7
ENST00000260733.7 |
TACC2
|
transforming acidic coiled-coil containing protein 2 |
| chr19_+_35139440 | 0.52 |
ENST00000455515.6
|
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr1_-_225941212 | 0.52 |
ENST00000366820.10
|
LEFTY2
|
left-right determination factor 2 |
| chr19_+_35138993 | 0.52 |
ENST00000612146.4
ENST00000589209.5 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chr17_+_7355123 | 0.52 |
ENST00000389982.8
ENST00000576060.6 ENST00000330767.4 |
TMEM95
|
transmembrane protein 95 |
| chr9_-_128067310 | 0.52 |
ENST00000373078.5
|
NAIF1
|
nuclear apoptosis inducing factor 1 |
| chr3_+_38039199 | 0.51 |
ENST00000346219.7
ENST00000308059.11 |
DLEC1
|
DLEC1 cilia and flagella associated protein |
| chr5_-_161546970 | 0.51 |
ENST00000675303.1
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr5_-_161546671 | 0.50 |
ENST00000517547.5
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr1_-_225941383 | 0.48 |
ENST00000420304.6
|
LEFTY2
|
left-right determination factor 2 |
| chr1_-_153565535 | 0.48 |
ENST00000368707.5
|
S100A2
|
S100 calcium binding protein A2 |
| chr21_+_42653585 | 0.48 |
ENST00000291539.11
|
PDE9A
|
phosphodiesterase 9A |
| chr10_-_122845850 | 0.48 |
ENST00000392790.6
|
CUZD1
|
CUB and zona pellucida like domains 1 |
| chr20_-_49913693 | 0.48 |
ENST00000422556.1
|
SPATA2
|
spermatogenesis associated 2 |
| chr2_+_11746576 | 0.48 |
ENST00000256720.6
ENST00000674199.1 ENST00000441684.5 ENST00000423495.1 |
LPIN1
|
lipin 1 |
| chr17_-_1492660 | 0.48 |
ENST00000648651.1
|
MYO1C
|
myosin IC |
| chr7_-_100158679 | 0.47 |
ENST00000456769.5
ENST00000316937.8 |
TRAPPC14
|
trafficking protein particle complex 14 |
| chr18_+_58362467 | 0.45 |
ENST00000675101.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr12_-_120904337 | 0.45 |
ENST00000353487.7
|
SPPL3
|
signal peptide peptidase like 3 |
| chr1_-_161549793 | 0.44 |
ENST00000443193.6
|
FCGR3A
|
Fc fragment of IgG receptor IIIa |
| chr22_+_21015027 | 0.44 |
ENST00000413302.6
ENST00000401443.5 |
P2RX6
|
purinergic receptor P2X 6 |
| chr3_+_63967738 | 0.44 |
ENST00000484332.1
|
ATXN7
|
ataxin 7 |
| chr11_-_57322197 | 0.43 |
ENST00000532437.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1 |
| chr19_+_41219235 | 0.43 |
ENST00000359092.7
|
AXL
|
AXL receptor tyrosine kinase |
| chr14_+_58637934 | 0.43 |
ENST00000395153.8
|
DACT1
|
dishevelled binding antagonist of beta catenin 1 |
| chr17_-_76027212 | 0.42 |
ENST00000586740.1
|
EVPL
|
envoplakin |
| chrX_+_137566119 | 0.42 |
ENST00000287538.10
|
ZIC3
|
Zic family member 3 |
| chr5_-_143403297 | 0.42 |
ENST00000415690.6
|
NR3C1
|
nuclear receptor subfamily 3 group C member 1 |
| chr22_-_22508702 | 0.42 |
ENST00000626650.3
|
ZNF280B
|
zinc finger protein 280B |
| chr4_-_173530219 | 0.42 |
ENST00000359562.4
|
HAND2
|
heart and neural crest derivatives expressed 2 |
| chr7_-_113086815 | 0.42 |
ENST00000424100.2
|
GPR85
|
G protein-coupled receptor 85 |
| chr1_-_161549892 | 0.42 |
ENST00000426740.7
|
FCGR3A
|
Fc fragment of IgG receptor IIIa |
| chr22_-_38088915 | 0.41 |
ENST00000428572.1
|
BAIAP2L2
|
BAR/IMD domain containing adaptor protein 2 like 2 |
| chr2_+_240625237 | 0.41 |
ENST00000407714.1
|
GPR35
|
G protein-coupled receptor 35 |
| chr2_+_71068636 | 0.40 |
ENST00000244204.11
ENST00000533981.5 |
NAGK
|
N-acetylglucosamine kinase |
| chr5_-_59356962 | 0.40 |
ENST00000405755.6
|
PDE4D
|
phosphodiesterase 4D |
| chr2_+_71068603 | 0.39 |
ENST00000443938.6
|
NAGK
|
N-acetylglucosamine kinase |
| chr12_-_52652207 | 0.38 |
ENST00000309680.4
|
KRT2
|
keratin 2 |
| chr19_+_35138778 | 0.37 |
ENST00000351325.9
ENST00000586871.5 ENST00000592174.1 |
FXYD1
ENSG00000221857.7
|
FXYD domain containing ion transport regulator 1 novel transcript |
| chr14_-_68979076 | 0.37 |
ENST00000538545.6
ENST00000684639.1 |
ACTN1
|
actinin alpha 1 |
| chr12_-_57772087 | 0.37 |
ENST00000324871.12
ENST00000257848.7 |
METTL1
|
methyltransferase like 1 |
| chr2_+_202073282 | 0.37 |
ENST00000459709.5
|
KIAA2012
|
KIAA2012 |
| chr12_-_119803383 | 0.36 |
ENST00000392520.2
ENST00000678677.1 ENST00000679249.1 ENST00000676849.1 |
CIT
|
citron rho-interacting serine/threonine kinase |
| chr14_-_68937942 | 0.36 |
ENST00000684182.1
|
ACTN1
|
actinin alpha 1 |
| chr12_-_53232182 | 0.36 |
ENST00000425354.7
ENST00000546717.1 ENST00000394426.5 |
RARG
|
retinoic acid receptor gamma |
| chr1_-_204359885 | 0.36 |
ENST00000414478.1
ENST00000272203.8 |
PLEKHA6
|
pleckstrin homology domain containing A6 |
| chr14_-_68979251 | 0.36 |
ENST00000438964.6
ENST00000679147.1 |
ACTN1
|
actinin alpha 1 |
| chr6_+_150683593 | 0.35 |
ENST00000644968.1
|
PLEKHG1
|
pleckstrin homology and RhoGEF domain containing G1 |
| chr3_-_50503597 | 0.35 |
ENST00000266039.7
ENST00000424201.7 |
CACNA2D2
|
calcium voltage-gated channel auxiliary subunit alpha2delta 2 |
| chr22_+_25199856 | 0.35 |
ENST00000215855.7
ENST00000404334.1 |
CRYBB3
|
crystallin beta B3 |
| chr3_-_52409783 | 0.35 |
ENST00000470173.1
ENST00000296288.9 ENST00000460680.6 |
BAP1
|
BRCA1 associated protein 1 |
| chr6_-_31112566 | 0.35 |
ENST00000259870.4
|
C6orf15
|
chromosome 6 open reading frame 15 |
| chr2_+_202073249 | 0.34 |
ENST00000498697.3
|
KIAA2012
|
KIAA2012 |
| chr1_-_207032749 | 0.34 |
ENST00000359470.6
ENST00000461135.2 |
C1orf116
|
chromosome 1 open reading frame 116 |
| chr17_-_82648428 | 0.34 |
ENST00000392325.9
|
WDR45B
|
WD repeat domain 45B |
| chr5_-_179806830 | 0.34 |
ENST00000292591.12
|
MGAT4B
|
alpha-1,3-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase B |
| chrX_+_70455093 | 0.33 |
ENST00000542398.1
|
DLG3
|
discs large MAGUK scaffold protein 3 |
| chr17_-_28951285 | 0.33 |
ENST00000577226.5
|
PHF12
|
PHD finger protein 12 |
| chr12_+_57772648 | 0.33 |
ENST00000333012.5
|
EEF1AKMT3
|
EEF1A lysine methyltransferase 3 |
| chr8_-_27992624 | 0.32 |
ENST00000524352.5
|
SCARA5
|
scavenger receptor class A member 5 |
| chr8_+_54457927 | 0.32 |
ENST00000297316.5
|
SOX17
|
SRY-box transcription factor 17 |
| chr1_+_193059587 | 0.32 |
ENST00000400968.7
ENST00000415442.2 ENST00000506303.1 |
RO60
|
Ro60, Y RNA binding protein |
| chr12_+_7789393 | 0.31 |
ENST00000229307.9
|
NANOG
|
Nanog homeobox |
| chr19_+_38930916 | 0.31 |
ENST00000308018.9
ENST00000407800.2 ENST00000402029.3 |
MRPS12
|
mitochondrial ribosomal protein S12 |
| chr22_+_20131781 | 0.31 |
ENST00000334554.12
ENST00000320602.11 ENST00000405930.3 |
ZDHHC8
|
zinc finger DHHC-type palmitoyltransferase 8 |
| chr15_+_60004305 | 0.31 |
ENST00000396057.6
|
FOXB1
|
forkhead box B1 |
| chr11_-_86069043 | 0.31 |
ENST00000532317.5
ENST00000528256.1 ENST00000393346.8 ENST00000526033.5 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr19_-_6481769 | 0.30 |
ENST00000381480.7
ENST00000543576.5 ENST00000590173.5 |
DENND1C
|
DENN domain containing 1C |
| chr22_-_42857194 | 0.30 |
ENST00000437119.6
ENST00000454099.5 ENST00000263245.10 |
ARFGAP3
|
ADP ribosylation factor GTPase activating protein 3 |
| chr12_+_57772587 | 0.29 |
ENST00000300209.13
|
EEF1AKMT3
|
EEF1A lysine methyltransferase 3 |
| chr3_-_48188356 | 0.29 |
ENST00000351231.7
ENST00000437972.1 ENST00000302506.8 |
CDC25A
|
cell division cycle 25A |
| chr2_+_11724333 | 0.29 |
ENST00000425416.6
ENST00000396097.5 |
LPIN1
|
lipin 1 |
| chr12_+_52079700 | 0.29 |
ENST00000546390.2
|
SMIM41
|
small integral membrane protein 41 |
| chr12_-_110445540 | 0.28 |
ENST00000547365.1
|
ARPC3
|
actin related protein 2/3 complex subunit 3 |
| chr5_-_141618914 | 0.28 |
ENST00000518047.5
|
DIAPH1
|
diaphanous related formin 1 |
| chr3_-_122416035 | 0.28 |
ENST00000330689.6
|
WDR5B
|
WD repeat domain 5B |
| chr11_-_78188588 | 0.28 |
ENST00000526208.5
ENST00000529350.1 ENST00000530018.5 ENST00000528776.1 ENST00000340067.4 |
KCTD21
|
potassium channel tetramerization domain containing 21 |
| chr1_+_193059448 | 0.28 |
ENST00000432079.5
|
RO60
|
Ro60, Y RNA binding protein |
| chr15_+_89690804 | 0.28 |
ENST00000268130.12
ENST00000560294.5 ENST00000558000.1 |
WDR93
|
WD repeat domain 93 |
| chr12_+_123633739 | 0.28 |
ENST00000618160.4
|
GTF2H3
|
general transcription factor IIH subunit 3 |
| chr2_-_20012661 | 0.28 |
ENST00000421259.2
ENST00000407540.8 |
MATN3
|
matrilin 3 |
| chr19_+_41219177 | 0.28 |
ENST00000301178.9
|
AXL
|
AXL receptor tyrosine kinase |
| chr19_-_12551426 | 0.27 |
ENST00000416136.1
ENST00000339282.12 ENST00000596193.1 ENST00000428311.1 |
ZNF564
ENSG00000196826.7
|
zinc finger protein 564 novel zinc finger protein |
| chr19_-_12294819 | 0.27 |
ENST00000355684.6
ENST00000356109.10 |
ZNF44
|
zinc finger protein 44 |
| chr11_+_100687279 | 0.27 |
ENST00000298815.13
|
ARHGAP42
|
Rho GTPase activating protein 42 |
| chr1_-_156248038 | 0.27 |
ENST00000470198.5
ENST00000292291.10 ENST00000356983.7 |
PAQR6
|
progestin and adipoQ receptor family member 6 |
| chr18_-_48950960 | 0.27 |
ENST00000262158.8
|
SMAD7
|
SMAD family member 7 |
| chr18_+_23949847 | 0.26 |
ENST00000588004.1
|
LAMA3
|
laminin subunit alpha 3 |
| chr7_-_122995700 | 0.26 |
ENST00000249284.3
|
TAS2R16
|
taste 2 receptor member 16 |
| chr2_+_231781669 | 0.26 |
ENST00000410024.5
ENST00000611614.4 ENST00000409295.5 ENST00000409091.5 |
COPS7B
|
COP9 signalosome subunit 7B |
| chr11_+_111245725 | 0.26 |
ENST00000280325.7
|
C11orf53
|
chromosome 11 open reading frame 53 |
| chr5_-_161546708 | 0.26 |
ENST00000393959.6
|
GABRB2
|
gamma-aminobutyric acid type A receptor subunit beta2 |
| chr11_-_128867268 | 0.26 |
ENST00000392665.6
ENST00000392666.6 |
KCNJ1
|
potassium inwardly rectifying channel subfamily J member 1 |
| chr17_-_1628341 | 0.26 |
ENST00000571650.5
|
SLC43A2
|
solute carrier family 43 member 2 |
| chr1_+_151540299 | 0.26 |
ENST00000392712.7
ENST00000368848.6 ENST00000368849.8 ENST00000353024.4 |
TUFT1
|
tuftelin 1 |
| chr1_+_110150480 | 0.25 |
ENST00000331565.5
|
SLC6A17
|
solute carrier family 6 member 17 |
| chr11_-_128867364 | 0.25 |
ENST00000440599.6
ENST00000324036.7 |
KCNJ1
|
potassium inwardly rectifying channel subfamily J member 1 |
| chr5_-_141619049 | 0.25 |
ENST00000647433.1
ENST00000253811.10 ENST00000389057.9 ENST00000398557.8 |
DIAPH1
|
diaphanous related formin 1 |
| chr5_+_176238365 | 0.25 |
ENST00000341199.10
ENST00000430704.6 ENST00000443967.5 ENST00000429602.7 |
SIMC1
|
SUMO interacting motifs containing 1 |
| chr12_+_123633819 | 0.25 |
ENST00000539994.5
ENST00000538845.5 ENST00000228955.11 ENST00000543341.7 ENST00000536375.5 |
GTF2H3
|
general transcription factor IIH subunit 3 |
| chr1_+_26863140 | 0.25 |
ENST00000339276.6
|
SFN
|
stratifin |
| chr5_-_141618957 | 0.25 |
ENST00000389054.8
|
DIAPH1
|
diaphanous related formin 1 |
| chr8_-_131040211 | 0.24 |
ENST00000377928.7
|
ADCY8
|
adenylate cyclase 8 |
| chr16_-_81096163 | 0.24 |
ENST00000566566.2
ENST00000569885.6 ENST00000561801.2 ENST00000639689.1 ENST00000638948.1 ENST00000564536.2 ENST00000638192.1 ENST00000640345.1 |
GCSH
ENSG00000260643.2
ENSG00000284512.1
|
glycine cleavage system protein H novel protein novel protein |
| chr3_-_50567711 | 0.23 |
ENST00000357203.8
|
C3orf18
|
chromosome 3 open reading frame 18 |
| chr1_-_156248013 | 0.23 |
ENST00000368270.2
|
PAQR6
|
progestin and adipoQ receptor family member 6 |
| chr1_+_116111395 | 0.23 |
ENST00000684484.1
ENST00000369500.4 |
MAB21L3
|
mab-21 like 3 |
| chr5_+_96663010 | 0.23 |
ENST00000506811.5
ENST00000514055.5 ENST00000508608.6 |
CAST
|
calpastatin |
| chr3_-_50567646 | 0.23 |
ENST00000426034.5
ENST00000441239.5 |
C3orf18
|
chromosome 3 open reading frame 18 |
| chr3_+_184362991 | 0.23 |
ENST00000430783.5
|
POLR2H
|
RNA polymerase II, I and III subunit H |
| chr1_+_204870831 | 0.23 |
ENST00000404076.5
ENST00000539706.6 |
NFASC
|
neurofascin |
| chr7_+_151956379 | 0.23 |
ENST00000431418.6
|
GALNTL5
|
polypeptide N-acetylgalactosaminyltransferase like 5 |
| chr5_-_177311882 | 0.22 |
ENST00000513169.1
ENST00000423571.6 ENST00000502529.1 ENST00000427908.6 |
MXD3
|
MAX dimerization protein 3 |
| chr10_-_70785371 | 0.22 |
ENST00000456372.4
ENST00000299290.5 |
TBATA
|
thymus, brain and testes associated |
| chr11_-_119423162 | 0.22 |
ENST00000284240.10
ENST00000524970.5 |
THY1
|
Thy-1 cell surface antigen |
| chr7_+_151956440 | 0.22 |
ENST00000392800.7
ENST00000616416.4 |
GALNTL5
|
polypeptide N-acetylgalactosaminyltransferase like 5 |
| chr19_+_18612848 | 0.22 |
ENST00000262817.8
|
TMEM59L
|
transmembrane protein 59 like |
| chr8_-_131040890 | 0.22 |
ENST00000286355.10
|
ADCY8
|
adenylate cyclase 8 |
| chr9_+_68780029 | 0.21 |
ENST00000394264.7
|
PABIR1
|
PP2A Aalpha (PPP2R1A) and B55A (PPP2R2A) interacting phosphatase regulator 1 |
| chr7_-_140479476 | 0.21 |
ENST00000443720.6
ENST00000255977.7 |
MKRN1
|
makorin ring finger protein 1 |
| chr12_+_53097656 | 0.21 |
ENST00000301464.4
|
IGFBP6
|
insulin like growth factor binding protein 6 |
| chr2_+_233251694 | 0.21 |
ENST00000417017.5
ENST00000392020.8 ENST00000392018.1 |
ATG16L1
|
autophagy related 16 like 1 |
| chr12_-_48004467 | 0.21 |
ENST00000380518.8
|
COL2A1
|
collagen type II alpha 1 chain |
| chr2_+_233251571 | 0.21 |
ENST00000347464.9
ENST00000444735.5 ENST00000373525.9 ENST00000392017.9 ENST00000419681.5 |
ATG16L1
|
autophagy related 16 like 1 |
| chr5_-_140557414 | 0.21 |
ENST00000336283.9
|
SRA1
|
steroid receptor RNA activator 1 |
| chr17_-_28951443 | 0.20 |
ENST00000268756.7
ENST00000332830.9 ENST00000584685.1 |
PHF12
|
PHD finger protein 12 |
| chr22_+_31160171 | 0.20 |
ENST00000326132.11
ENST00000426256.6 ENST00000266252.8 |
RNF185
|
ring finger protein 185 |
| chr1_-_161132423 | 0.20 |
ENST00000458050.6
|
DEDD
|
death effector domain containing |
| chr11_+_78188871 | 0.20 |
ENST00000528910.5
ENST00000529308.6 |
USP35
|
ubiquitin specific peptidase 35 |
| chr19_-_4065732 | 0.20 |
ENST00000601588.1
|
ZBTB7A
|
zinc finger and BTB domain containing 7A |
| chr22_+_22734577 | 0.19 |
ENST00000390310.3
|
IGLV2-18
|
immunoglobulin lambda variable 2-18 |
| chr12_+_14419136 | 0.19 |
ENST00000545769.1
ENST00000428217.6 ENST00000396279.2 ENST00000542514.5 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr20_+_45406035 | 0.19 |
ENST00000372723.7
ENST00000372722.7 |
DBNDD2
|
dysbindin domain containing 2 |
| chr6_-_18155053 | 0.19 |
ENST00000309983.5
|
TPMT
|
thiopurine S-methyltransferase |
| chr4_-_56435581 | 0.19 |
ENST00000264220.6
|
PPAT
|
phosphoribosyl pyrophosphate amidotransferase |
| chr8_-_143986425 | 0.19 |
ENST00000313059.9
ENST00000524918.5 ENST00000313028.12 ENST00000525773.5 |
PARP10
|
poly(ADP-ribose) polymerase family member 10 |
| chr13_-_36131286 | 0.19 |
ENST00000255448.8
ENST00000379892.4 |
DCLK1
|
doublecortin like kinase 1 |
| chrX_-_50200988 | 0.19 |
ENST00000358526.7
|
AKAP4
|
A-kinase anchoring protein 4 |
| chr16_+_30699155 | 0.19 |
ENST00000262518.9
|
SRCAP
|
Snf2 related CREBBP activator protein |
| chr5_-_140043069 | 0.18 |
ENST00000289409.8
ENST00000358522.7 ENST00000289422.11 ENST00000541337.5 ENST00000361474.6 |
NRG2
|
neuregulin 2 |
| chr8_+_116766497 | 0.18 |
ENST00000517814.1
ENST00000309822.7 ENST00000517820.1 |
UTP23
|
UTP23 small subunit processome component |
| chr22_-_38755458 | 0.18 |
ENST00000405510.5
ENST00000433561.5 |
SUN2
|
Sad1 and UNC84 domain containing 2 |
| chr5_+_163437569 | 0.18 |
ENST00000512163.5
ENST00000393929.5 ENST00000510097.5 ENST00000340828.7 ENST00000511490.4 ENST00000510664.5 |
CCNG1
|
cyclin G1 |
| chr7_+_43926419 | 0.18 |
ENST00000222402.8
ENST00000446008.5 |
UBE2D4
|
ubiquitin conjugating enzyme E2 D4 (putative) |
| chr10_-_126388455 | 0.18 |
ENST00000368679.8
ENST00000368676.8 ENST00000448723.2 |
ADAM12
|
ADAM metallopeptidase domain 12 |
| chr7_-_128405930 | 0.18 |
ENST00000470772.5
ENST00000480861.5 ENST00000496200.5 |
IMPDH1
|
inosine monophosphate dehydrogenase 1 |
| chrX_-_107717054 | 0.18 |
ENST00000503515.1
ENST00000372397.6 |
TSC22D3
|
TSC22 domain family member 3 |
| chr16_+_22008083 | 0.17 |
ENST00000542527.7
ENST00000569656.5 ENST00000562695.1 |
MOSMO
|
modulator of smoothened |
| chr9_+_85941121 | 0.17 |
ENST00000361671.10
|
NAA35
|
N-alpha-acetyltransferase 35, NatC auxiliary subunit |
| chr20_+_13221257 | 0.17 |
ENST00000262487.5
|
ISM1
|
isthmin 1 |
| chr4_-_173334385 | 0.17 |
ENST00000446922.6
|
HMGB2
|
high mobility group box 2 |
| chr17_+_16415553 | 0.17 |
ENST00000338560.12
|
TRPV2
|
transient receptor potential cation channel subfamily V member 2 |
| chr19_+_23117018 | 0.17 |
ENST00000597761.7
|
ZNF730
|
zinc finger protein 730 |
| chr11_-_33161461 | 0.17 |
ENST00000323959.9
ENST00000431742.2 ENST00000524827.6 |
CSTF3
|
cleavage stimulation factor subunit 3 |
| chr16_+_89508367 | 0.17 |
ENST00000643649.1
ENST00000643307.1 ENST00000566371.6 ENST00000645897.1 ENST00000644781.1 ENST00000645063.1 ENST00000645818.2 ENST00000646716.1 ENST00000268704.7 ENST00000341316.6 |
SPG7
|
SPG7 matrix AAA peptidase subunit, paraplegin |
| chr5_+_150671588 | 0.17 |
ENST00000523553.1
|
MYOZ3
|
myozenin 3 |
| chr1_-_161038907 | 0.17 |
ENST00000318289.14
ENST00000368023.7 ENST00000423014.3 ENST00000368024.5 |
TSTD1
|
thiosulfate sulfurtransferase like domain containing 1 |
| chr1_+_50106265 | 0.16 |
ENST00000357083.8
|
ELAVL4
|
ELAV like RNA binding protein 4 |
| chr4_+_82430563 | 0.16 |
ENST00000273920.8
|
ENOPH1
|
enolase-phosphatase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.7 | GO:0021564 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) vagus nerve development(GO:0021564) |
| 0.4 | 1.1 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.3 | 1.7 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.3 | 0.3 | GO:0061373 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) |
| 0.2 | 1.0 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.2 | 1.4 | GO:0010731 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.2 | 2.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.2 | 0.6 | GO:0070256 | germinal center B cell differentiation(GO:0002314) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) xanthine metabolic process(GO:0046110) negative regulation of mucus secretion(GO:0070256) |
| 0.1 | 0.8 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.1 | 0.8 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.1 | 0.8 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 0.3 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) cardiac cell fate determination(GO:0060913) |
| 0.1 | 0.9 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 | 0.7 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 0.1 | 0.8 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 0.6 | GO:0060356 | leucine import(GO:0060356) |
| 0.1 | 0.4 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.1 | 0.4 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.1 | 0.4 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 | 0.4 | GO:0061032 | cardiac right ventricle formation(GO:0003219) visceral serous pericardium development(GO:0061032) |
| 0.1 | 1.4 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.1 | 2.0 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.1 | 0.5 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 1.2 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.1 | 0.2 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 | 0.2 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 0.4 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.5 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.3 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 1.1 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 0.5 | GO:1902962 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.1 | 0.3 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 1.8 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.5 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.4 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 0.4 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 0.4 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 0.2 | GO:1902232 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) regulation of positive thymic T cell selection(GO:1902232) |
| 0.0 | 0.3 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.2 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.3 | GO:0015820 | leucine transport(GO:0015820) |
| 0.0 | 0.4 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0010902 | positive regulation of very-low-density lipoprotein particle remodeling(GO:0010902) |
| 0.0 | 0.3 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.2 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.4 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 0.1 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.0 | 0.2 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 2.6 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.2 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.3 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.3 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 1.1 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0042851 | L-alanine metabolic process(GO:0042851) L-alanine catabolic process(GO:0042853) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0097694 | regulation of telomere maintenance via semi-conservative replication(GO:0032213) negative regulation of telomere maintenance via semi-conservative replication(GO:0032214) establishment of RNA localization to telomere(GO:0097694) establishment of macromolecular complex localization to telomere(GO:0097695) |
| 0.0 | 1.1 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:1904349 | positive regulation of eating behavior(GO:1904000) regulation of small intestine smooth muscle contraction(GO:1904347) positive regulation of small intestine smooth muscle contraction(GO:1904349) small intestine smooth muscle contraction(GO:1990770) |
| 0.0 | 0.3 | GO:0034380 | high-density lipoprotein particle assembly(GO:0034380) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.2 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0003342 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.6 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.1 | GO:0043366 | beta selection(GO:0043366) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.1 | GO:1990086 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.1 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 1.3 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.3 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.5 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.2 | GO:0045654 | DNA topological change(GO:0006265) positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.5 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.4 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.3 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.0 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.4 | GO:0061045 | negative regulation of wound healing(GO:0061045) |
| 0.0 | 1.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.0 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.1 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.0 | 0.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.8 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.9 | GO:0032945 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of lymphocyte proliferation(GO:0050672) |
| 0.0 | 0.1 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0098560 | cytoplasmic side of early endosome membrane(GO:0098559) cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 0.5 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.1 | 3.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.5 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.5 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.3 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 1.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.3 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.1 | 0.4 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.1 | 1.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 1.0 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.2 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 1.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.3 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.2 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.3 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.7 | GO:0033643 | host cell part(GO:0033643) |
| 0.0 | 0.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.1 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.8 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.5 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.5 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.6 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.2 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.3 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 1.1 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.2 | 0.8 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.2 | 1.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.2 | 0.5 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 1.3 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.1 | 0.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.3 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.1 | 1.7 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 0.4 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 1.3 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.5 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.2 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.1 | 0.2 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 0.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 0.2 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.1 | 0.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.3 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 1.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.8 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.3 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.1 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.0 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 1.5 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.2 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 1.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.7 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.4 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 1.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.3 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 1.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.5 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 1.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.6 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.5 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 1.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.0 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 1.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.4 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 2.0 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.0 | GO:0097259 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.1 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 1.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 2.5 | GO:0008201 | heparin binding(GO:0008201) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 2.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 2.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.7 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.8 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.7 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.7 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 1.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.3 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.5 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |