Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
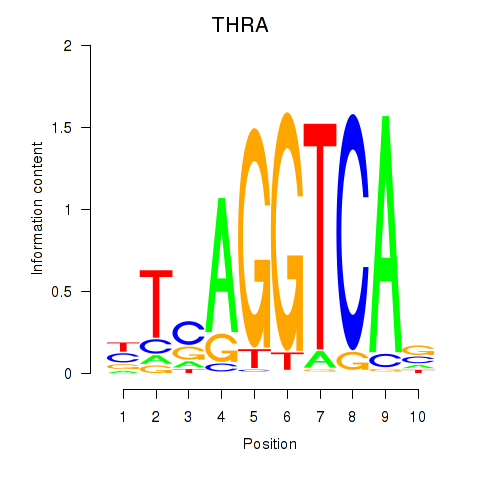
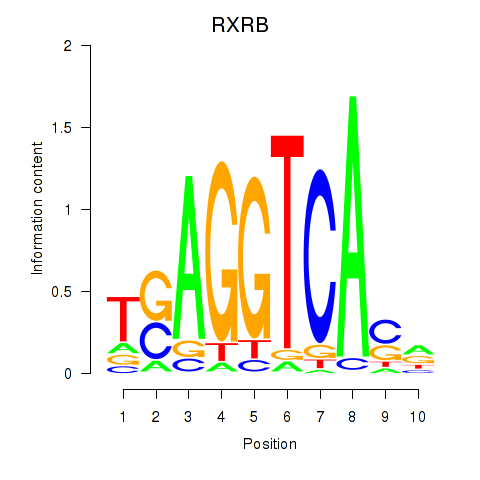
Results for THRA_RXRB
Z-value: 0.79
Transcription factors associated with THRA_RXRB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
THRA
|
ENSG00000126351.13 | THRA |
|
RXRB
|
ENSG00000204231.11 | RXRB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| THRA | hg38_v1_chr17_+_40062810_40062901 | 0.33 | 1.1e-01 | Click! |
| RXRB | hg38_v1_chr6_-_33200614_33200680 | 0.00 | 1.0e+00 | Click! |
Activity profile of THRA_RXRB motif
Sorted Z-values of THRA_RXRB motif
Network of associatons between targets according to the STRING database.
First level regulatory network of THRA_RXRB
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_62248591 | 1.63 |
ENST00000519049.6
|
NKAIN3
|
sodium/potassium transporting ATPase interacting 3 |
| chr4_-_148444674 | 1.09 |
ENST00000344721.8
|
NR3C2
|
nuclear receptor subfamily 3 group C member 2 |
| chr19_+_1248553 | 0.99 |
ENST00000586757.5
ENST00000300952.6 ENST00000682408.1 |
MIDN
|
midnolin |
| chr9_-_33402551 | 0.79 |
ENST00000297988.6
ENST00000624075.3 ENST00000625032.1 ENST00000625109.3 |
AQP7
|
aquaporin 7 |
| chr2_-_55232158 | 0.77 |
ENST00000407122.5
ENST00000401408.6 |
CLHC1
|
clathrin heavy chain linker domain containing 1 |
| chr12_+_78863962 | 0.76 |
ENST00000393240.7
|
SYT1
|
synaptotagmin 1 |
| chr9_+_27109393 | 0.74 |
ENST00000406359.8
|
TEK
|
TEK receptor tyrosine kinase |
| chr14_+_88385643 | 0.72 |
ENST00000393545.9
ENST00000356583.9 ENST00000555401.5 ENST00000553885.5 |
SPATA7
|
spermatogenesis associated 7 |
| chr14_+_88385714 | 0.72 |
ENST00000045347.11
|
SPATA7
|
spermatogenesis associated 7 |
| chr11_+_7513966 | 0.67 |
ENST00000299492.9
|
PPFIBP2
|
PPFIA binding protein 2 |
| chr1_+_163069353 | 0.66 |
ENST00000531057.5
ENST00000527809.5 ENST00000367908.8 ENST00000367909.11 |
RGS4
|
regulator of G protein signaling 4 |
| chr1_+_32362537 | 0.63 |
ENST00000373534.4
|
TSSK3
|
testis specific serine kinase 3 |
| chr11_-_27700472 | 0.61 |
ENST00000418212.5
ENST00000533246.5 |
BDNF
|
brain derived neurotrophic factor |
| chr11_-_27700447 | 0.60 |
ENST00000356660.9
|
BDNF
|
brain derived neurotrophic factor |
| chr9_-_33402449 | 0.60 |
ENST00000377425.8
|
AQP7
|
aquaporin 7 |
| chr2_-_182522703 | 0.58 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A |
| chr19_-_51723968 | 0.57 |
ENST00000222115.5
ENST00000540069.7 |
HAS1
|
hyaluronan synthase 1 |
| chr1_+_160177386 | 0.56 |
ENST00000470705.1
|
ATP1A4
|
ATPase Na+/K+ transporting subunit alpha 4 |
| chr22_+_22822658 | 0.52 |
ENST00000620395.2
|
IGLV2-8
|
immunoglobulin lambda variable 2-8 |
| chr12_-_95116967 | 0.51 |
ENST00000551521.5
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr14_-_105588322 | 0.51 |
ENST00000497872.4
ENST00000390539.2 |
IGHA2
|
immunoglobulin heavy constant alpha 2 (A2m marker) |
| chr1_+_185734362 | 0.51 |
ENST00000271588.9
|
HMCN1
|
hemicentin 1 |
| chr5_-_163460067 | 0.51 |
ENST00000302764.9
|
NUDCD2
|
NudC domain containing 2 |
| chr14_-_76826229 | 0.50 |
ENST00000557497.1
|
ANGEL1
|
angel homolog 1 |
| chr17_-_41060109 | 0.49 |
ENST00000391418.3
|
KRTAP2-3
|
keratin associated protein 2-3 |
| chr12_+_57583101 | 0.47 |
ENST00000674858.1
ENST00000675433.1 ENST00000674980.1 |
KIF5A
|
kinesin family member 5A |
| chr12_-_52601458 | 0.43 |
ENST00000537672.6
ENST00000293745.7 |
KRT72
|
keratin 72 |
| chrX_-_133753681 | 0.43 |
ENST00000406757.2
|
GPC3
|
glypican 3 |
| chr5_-_163460048 | 0.42 |
ENST00000517501.1
|
NUDCD2
|
NudC domain containing 2 |
| chr12_-_52601507 | 0.42 |
ENST00000354310.4
|
KRT72
|
keratin 72 |
| chr15_-_79923647 | 0.42 |
ENST00000485386.1
ENST00000479961.1 ENST00000494999.1 |
ST20
ST20-MTHFS
|
suppressor of tumorigenicity 20 ST20-MTHFS readthrough |
| chr21_+_44353607 | 0.41 |
ENST00000397928.6
|
TRPM2
|
transient receptor potential cation channel subfamily M member 2 |
| chr3_+_37861926 | 0.40 |
ENST00000443503.6
|
CTDSPL
|
CTD small phosphatase like |
| chr22_-_21227637 | 0.40 |
ENST00000401924.5
|
GGT2
|
gamma-glutamyltransferase 2 |
| chr22_+_22594528 | 0.40 |
ENST00000390303.3
|
IGLV3-32
|
immunoglobulin lambda variable 3-32 (non-functional) |
| chr10_-_70376779 | 0.39 |
ENST00000395011.5
|
LRRC20
|
leucine rich repeat containing 20 |
| chr14_+_22495890 | 0.38 |
ENST00000390494.1
|
TRAJ43
|
T cell receptor alpha joining 43 |
| chr22_+_22922594 | 0.38 |
ENST00000390331.3
|
IGLC7
|
immunoglobulin lambda constant 7 |
| chr22_+_22792485 | 0.37 |
ENST00000390314.2
|
IGLV2-11
|
immunoglobulin lambda variable 2-11 |
| chr7_+_121328991 | 0.37 |
ENST00000222462.3
|
WNT16
|
Wnt family member 16 |
| chr22_+_22758698 | 0.36 |
ENST00000390312.2
|
IGLV2-14
|
immunoglobulin lambda variable 2-14 |
| chr3_+_184337591 | 0.36 |
ENST00000383847.7
|
FAM131A
|
family with sequence similarity 131 member A |
| chr17_+_45844875 | 0.36 |
ENST00000329196.7
|
SPPL2C
|
signal peptide peptidase like 2C |
| chr10_-_96357155 | 0.35 |
ENST00000536387.5
|
OPALIN
|
oligodendrocytic myelin paranodal and inner loop protein |
| chr22_+_30881674 | 0.35 |
ENST00000454145.5
ENST00000453621.5 ENST00000431368.5 ENST00000535268.5 |
OSBP2
|
oxysterol binding protein 2 |
| chr6_+_139135063 | 0.34 |
ENST00000367658.3
|
HECA
|
hdc homolog, cell cycle regulator |
| chr21_+_44353633 | 0.34 |
ENST00000397932.6
ENST00000300481.13 |
TRPM2
|
transient receptor potential cation channel subfamily M member 2 |
| chr22_+_28742024 | 0.33 |
ENST00000216027.8
ENST00000398941.6 |
HSCB
|
HscB mitochondrial iron-sulfur cluster cochaperone |
| chr12_+_102120172 | 0.33 |
ENST00000327680.7
ENST00000541394.5 ENST00000543784.5 |
PARPBP
|
PARP1 binding protein |
| chr11_+_64340191 | 0.33 |
ENST00000356786.10
|
CCDC88B
|
coiled-coil domain containing 88B |
| chr2_-_74552616 | 0.33 |
ENST00000409249.5
|
LOXL3
|
lysyl oxidase like 3 |
| chr7_-_92246045 | 0.33 |
ENST00000394507.5
ENST00000458177.6 ENST00000340022.6 ENST00000444960.5 |
KRIT1
|
KRIT1 ankyrin repeat containing |
| chr12_-_95790755 | 0.32 |
ENST00000343702.9
ENST00000344911.8 |
NTN4
|
netrin 4 |
| chr2_-_182522556 | 0.31 |
ENST00000435564.5
|
PDE1A
|
phosphodiesterase 1A |
| chrX_-_111410420 | 0.31 |
ENST00000371993.7
ENST00000680476.1 |
DCX
|
doublecortin |
| chr13_-_33185994 | 0.31 |
ENST00000255486.8
|
STARD13
|
StAR related lipid transfer domain containing 13 |
| chr22_+_22327298 | 0.30 |
ENST00000390291.2
|
IGLV1-50
|
immunoglobulin lambda variable 1-50 (non-functional) |
| chr7_-_92245795 | 0.30 |
ENST00000412043.6
ENST00000430102.5 ENST00000425073.1 ENST00000394505.7 ENST00000394503.6 ENST00000454017.5 ENST00000440209.5 ENST00000413688.5 ENST00000452773.5 ENST00000433016.5 ENST00000422347.5 ENST00000458493.5 ENST00000425919.5 ENST00000650585.1 |
KRIT1
ENSG00000285953.1
|
KRIT1 ankyrin repeat containing novel protein |
| chr1_+_70411180 | 0.30 |
ENST00000411986.6
|
CTH
|
cystathionine gamma-lyase |
| chr14_+_74084947 | 0.30 |
ENST00000674221.1
ENST00000554938.2 |
LIN52
|
lin-52 DREAM MuvB core complex component |
| chr2_-_69643152 | 0.29 |
ENST00000606389.7
|
AAK1
|
AP2 associated kinase 1 |
| chr5_+_170861990 | 0.29 |
ENST00000523189.6
|
RANBP17
|
RAN binding protein 17 |
| chr22_-_37484505 | 0.29 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr1_-_150720842 | 0.28 |
ENST00000442853.5
ENST00000368995.8 ENST00000322343.11 ENST00000361824.7 |
HORMAD1
|
HORMA domain containing 1 |
| chr17_-_2025289 | 0.28 |
ENST00000331238.7
|
RTN4RL1
|
reticulon 4 receptor like 1 |
| chr17_+_46851580 | 0.27 |
ENST00000290015.7
ENST00000393461.2 |
WNT9B
|
Wnt family member 9B |
| chr12_-_111488538 | 0.27 |
ENST00000389154.7
|
ATXN2
|
ataxin 2 |
| chr8_-_18808837 | 0.27 |
ENST00000286485.12
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr9_-_101487120 | 0.27 |
ENST00000374848.8
|
PGAP4
|
post-GPI attachment to proteins GalNAc transferase 4 |
| chr14_-_105708627 | 0.27 |
ENST00000641837.1
ENST00000390547.3 |
IGHA1
|
immunoglobulin heavy constant alpha 1 |
| chr3_+_40477107 | 0.27 |
ENST00000314686.9
ENST00000447116.6 ENST00000429348.6 ENST00000432264.4 ENST00000456778.5 |
ZNF619
|
zinc finger protein 619 |
| chr3_-_50303565 | 0.26 |
ENST00000266031.8
ENST00000395143.6 ENST00000457214.6 ENST00000447605.2 ENST00000395144.7 ENST00000418723.1 |
HYAL1
|
hyaluronidase 1 |
| chr11_-_71928624 | 0.26 |
ENST00000533047.5
ENST00000529844.5 |
ENSG00000254469.9
|
novel protein similar to transient receptor potential cation channel, subfamily C, member 2 TRPC2 |
| chr13_-_75537805 | 0.26 |
ENST00000626103.1
ENST00000682242.1 ENST00000355801.4 |
COMMD6
|
COMM domain containing 6 |
| chr22_+_22162155 | 0.26 |
ENST00000390284.2
|
IGLV4-60
|
immunoglobulin lambda variable 4-60 |
| chr17_-_5234801 | 0.26 |
ENST00000571800.5
ENST00000574081.6 ENST00000399600.8 ENST00000574297.1 |
SCIMP
|
SLP adaptor and CSK interacting membrane protein |
| chr15_-_51622738 | 0.25 |
ENST00000560891.6
|
DMXL2
|
Dmx like 2 |
| chr3_-_49689053 | 0.25 |
ENST00000449682.2
|
MST1
|
macrophage stimulating 1 |
| chr1_+_70411241 | 0.25 |
ENST00000370938.8
ENST00000346806.2 |
CTH
|
cystathionine gamma-lyase |
| chr9_-_101487091 | 0.25 |
ENST00000374847.5
|
PGAP4
|
post-GPI attachment to proteins GalNAc transferase 4 |
| chr19_-_57974527 | 0.25 |
ENST00000314391.3
|
C19orf18
|
chromosome 19 open reading frame 18 |
| chr17_+_29043124 | 0.25 |
ENST00000323372.9
|
PIPOX
|
pipecolic acid and sarcosine oxidase |
| chr12_+_119593758 | 0.25 |
ENST00000426426.3
|
TMEM233
|
transmembrane protein 233 |
| chr20_-_37527862 | 0.25 |
ENST00000373537.7
ENST00000445723.5 ENST00000414080.1 |
BLCAP
|
BLCAP apoptosis inducing factor |
| chr1_+_28438104 | 0.24 |
ENST00000633167.1
ENST00000373836.4 |
PHACTR4
|
phosphatase and actin regulator 4 |
| chr15_-_34337462 | 0.24 |
ENST00000676379.1
|
SLC12A6
|
solute carrier family 12 member 6 |
| chr6_-_88963573 | 0.24 |
ENST00000369485.9
|
RNGTT
|
RNA guanylyltransferase and 5'-phosphatase |
| chr14_-_105601728 | 0.24 |
ENST00000641420.1
ENST00000390541.2 |
IGHE
|
immunoglobulin heavy constant epsilon |
| chr15_-_56733473 | 0.24 |
ENST00000558320.5
ENST00000267807.12 |
ZNF280D
|
zinc finger protein 280D |
| chr1_-_110606009 | 0.23 |
ENST00000640774.2
ENST00000638616.2 |
KCNA2
|
potassium voltage-gated channel subfamily A member 2 |
| chr15_-_45129857 | 0.23 |
ENST00000267803.8
ENST00000613425.4 ENST00000559014.5 ENST00000558851.1 ENST00000559988.5 ENST00000558996.5 ENST00000558422.5 ENST00000559226.5 ENST00000560572.6 ENST00000558326.5 ENST00000558377.5 ENST00000559644.5 |
DUOXA1
|
dual oxidase maturation factor 1 |
| chr22_+_22871478 | 0.23 |
ENST00000390318.2
|
IGLV4-3
|
immunoglobulin lambda variable 4-3 |
| chr4_+_89901979 | 0.23 |
ENST00000508372.1
|
MMRN1
|
multimerin 1 |
| chr3_+_12287859 | 0.23 |
ENST00000309576.11
ENST00000397015.7 |
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr1_+_178513103 | 0.23 |
ENST00000319416.7
ENST00000367643.7 ENST00000367642.3 ENST00000367641.7 ENST00000367639.1 |
TEX35
|
testis expressed 35 |
| chr17_-_60392333 | 0.23 |
ENST00000590133.5
|
USP32
|
ubiquitin specific peptidase 32 |
| chr19_+_19528861 | 0.23 |
ENST00000436027.9
|
YJEFN3
|
YjeF N-terminal domain containing 3 |
| chr3_+_12287962 | 0.22 |
ENST00000643197.2
ENST00000644622.2 |
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr3_+_10026409 | 0.22 |
ENST00000287647.7
ENST00000676013.1 ENST00000675286.1 ENST00000419585.5 |
FANCD2
|
FA complementation group D2 |
| chr3_+_12287899 | 0.22 |
ENST00000643888.2
|
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr12_+_56007484 | 0.22 |
ENST00000262032.9
|
IKZF4
|
IKAROS family zinc finger 4 |
| chr2_-_74553934 | 0.22 |
ENST00000264094.8
ENST00000393937.6 ENST00000409986.5 |
LOXL3
|
lysyl oxidase like 3 |
| chr19_+_19528901 | 0.22 |
ENST00000514277.6
|
YJEFN3
|
YjeF N-terminal domain containing 3 |
| chr10_-_118046574 | 0.21 |
ENST00000369199.5
|
RAB11FIP2
|
RAB11 family interacting protein 2 |
| chr17_+_60422483 | 0.21 |
ENST00000269127.5
|
C17orf64
|
chromosome 17 open reading frame 64 |
| chrX_-_10833643 | 0.21 |
ENST00000380785.5
ENST00000380787.5 |
MID1
|
midline 1 |
| chr1_-_173917281 | 0.21 |
ENST00000367698.4
|
SERPINC1
|
serpin family C member 1 |
| chr17_-_7179544 | 0.20 |
ENST00000619926.4
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr17_+_66964638 | 0.20 |
ENST00000262138.4
|
CACNG4
|
calcium voltage-gated channel auxiliary subunit gamma 4 |
| chr6_+_13272709 | 0.20 |
ENST00000379335.8
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr10_-_118046922 | 0.20 |
ENST00000355624.8
|
RAB11FIP2
|
RAB11 family interacting protein 2 |
| chr17_+_49969178 | 0.20 |
ENST00000240306.5
|
DLX4
|
distal-less homeobox 4 |
| chr11_-_77411883 | 0.19 |
ENST00000528203.5
ENST00000528592.5 ENST00000528633.1 ENST00000529248.5 |
PAK1
|
p21 (RAC1) activated kinase 1 |
| chr8_-_98942557 | 0.19 |
ENST00000523601.5
|
STK3
|
serine/threonine kinase 3 |
| chr1_+_154327737 | 0.19 |
ENST00000672630.1
|
ATP8B2
|
ATPase phospholipid transporting 8B2 |
| chr12_-_6556034 | 0.19 |
ENST00000619571.5
ENST00000336604.8 ENST00000396840.6 ENST00000356896.8 |
IFFO1
|
intermediate filament family orphan 1 |
| chr16_+_28863812 | 0.19 |
ENST00000684370.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr22_-_37188281 | 0.19 |
ENST00000397110.6
|
C1QTNF6
|
C1q and TNF related 6 |
| chr16_-_58295019 | 0.19 |
ENST00000567164.6
ENST00000219301.8 ENST00000569727.1 |
PRSS54
|
serine protease 54 |
| chr7_+_77122609 | 0.19 |
ENST00000285871.5
|
CCDC146
|
coiled-coil domain containing 146 |
| chr12_-_52553139 | 0.18 |
ENST00000267119.6
|
KRT71
|
keratin 71 |
| chr17_+_39738317 | 0.18 |
ENST00000394211.7
|
GRB7
|
growth factor receptor bound protein 7 |
| chr1_+_17308194 | 0.18 |
ENST00000375453.5
ENST00000375448.4 |
PADI4
|
peptidyl arginine deiminase 4 |
| chr19_-_37207074 | 0.18 |
ENST00000588873.1
|
ENSG00000267360.6
|
novel protein |
| chr7_-_151736304 | 0.18 |
ENST00000492843.6
|
PRKAG2
|
protein kinase AMP-activated non-catalytic subunit gamma 2 |
| chr2_+_172821575 | 0.18 |
ENST00000397087.7
|
RAPGEF4
|
Rap guanine nucleotide exchange factor 4 |
| chr5_+_148312416 | 0.18 |
ENST00000274565.5
|
SPINK7
|
serine peptidase inhibitor Kazal type 7 |
| chr7_+_143284930 | 0.18 |
ENST00000409244.5
ENST00000409541.5 ENST00000410004.1 ENST00000359333.8 |
TMEM139
|
transmembrane protein 139 |
| chrX_+_48786578 | 0.18 |
ENST00000376670.9
|
GATA1
|
GATA binding protein 1 |
| chr16_-_2329687 | 0.18 |
ENST00000567910.1
|
ABCA3
|
ATP binding cassette subfamily A member 3 |
| chr14_-_70188967 | 0.18 |
ENST00000381269.6
ENST00000357887.7 |
SLC8A3
|
solute carrier family 8 member A3 |
| chr15_-_82647503 | 0.18 |
ENST00000567678.1
ENST00000620182.4 |
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr11_-_59810750 | 0.18 |
ENST00000300151.5
|
MRPL16
|
mitochondrial ribosomal protein L16 |
| chr9_+_17579059 | 0.18 |
ENST00000380607.5
|
SH3GL2
|
SH3 domain containing GRB2 like 2, endophilin A1 |
| chr2_+_218880844 | 0.18 |
ENST00000258411.8
|
WNT10A
|
Wnt family member 10A |
| chr2_+_148021083 | 0.18 |
ENST00000642680.2
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr15_-_51622798 | 0.17 |
ENST00000251076.9
|
DMXL2
|
Dmx like 2 |
| chr12_+_65169546 | 0.17 |
ENST00000308330.3
|
LEMD3
|
LEM domain containing 3 |
| chrX_-_136780925 | 0.17 |
ENST00000250617.7
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor 6 |
| chr20_+_18145083 | 0.17 |
ENST00000489634.2
|
KAT14
|
lysine acetyltransferase 14 |
| chr8_+_102551583 | 0.17 |
ENST00000285402.4
|
ODF1
|
outer dense fiber of sperm tails 1 |
| chr2_+_148021001 | 0.17 |
ENST00000407073.5
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chrX_-_130165699 | 0.17 |
ENST00000676328.1
ENST00000675857.1 ENST00000675427.1 ENST00000675092.1 |
AIFM1
|
apoptosis inducing factor mitochondria associated 1 |
| chr5_+_113513674 | 0.17 |
ENST00000161863.9
ENST00000515883.5 |
YTHDC2
|
YTH domain containing 2 |
| chr14_+_22883220 | 0.17 |
ENST00000536884.1
ENST00000267396.9 |
REM2
|
RRAD and GEM like GTPase 2 |
| chr2_+_74513441 | 0.17 |
ENST00000621092.1
|
TLX2
|
T cell leukemia homeobox 2 |
| chr1_-_242449478 | 0.17 |
ENST00000427495.5
|
PLD5
|
phospholipase D family member 5 |
| chr7_+_94907584 | 0.17 |
ENST00000433360.6
ENST00000340694.8 ENST00000424654.5 |
PPP1R9A
|
protein phosphatase 1 regulatory subunit 9A |
| chr16_+_67666770 | 0.16 |
ENST00000403458.9
ENST00000602365.1 |
C16orf86
|
chromosome 16 open reading frame 86 |
| chr4_-_53591477 | 0.16 |
ENST00000263925.8
|
LNX1
|
ligand of numb-protein X 1 |
| chr2_+_61162185 | 0.16 |
ENST00000432605.3
ENST00000488469.4 |
C2orf74
|
chromosome 2 open reading frame 74 |
| chr1_-_155911340 | 0.16 |
ENST00000368323.8
|
RIT1
|
Ras like without CAAX 1 |
| chr14_-_70188937 | 0.16 |
ENST00000356921.7
|
SLC8A3
|
solute carrier family 8 member A3 |
| chr3_-_197298558 | 0.16 |
ENST00000656944.1
ENST00000346964.6 ENST00000448528.6 ENST00000655488.1 ENST00000357674.9 ENST00000667157.1 ENST00000661336.1 ENST00000654737.1 ENST00000659716.1 ENST00000657381.1 |
DLG1
|
discs large MAGUK scaffold protein 1 |
| chr9_+_27109200 | 0.15 |
ENST00000380036.10
|
TEK
|
TEK receptor tyrosine kinase |
| chr17_+_37491464 | 0.15 |
ENST00000613659.1
|
DUSP14
|
dual specificity phosphatase 14 |
| chr1_-_243843226 | 0.15 |
ENST00000336199.9
|
AKT3
|
AKT serine/threonine kinase 3 |
| chr5_+_163460623 | 0.15 |
ENST00000393915.9
ENST00000432118.6 |
HMMR
|
hyaluronan mediated motility receptor |
| chr3_+_12288838 | 0.15 |
ENST00000455517.6
ENST00000681982.1 |
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr16_-_67666692 | 0.15 |
ENST00000602644.5
ENST00000243878.9 |
ENKD1
|
enkurin domain containing 1 |
| chr12_+_102120240 | 0.15 |
ENST00000537257.5
ENST00000392911.6 |
PARPBP
|
PARP1 binding protein |
| chr2_+_230996115 | 0.15 |
ENST00000424440.5
ENST00000452881.5 ENST00000433428.6 ENST00000455816.1 ENST00000440792.5 ENST00000423134.1 |
SPATA3
|
spermatogenesis associated 3 |
| chr1_+_93079234 | 0.15 |
ENST00000540243.5
ENST00000545708.5 |
MTF2
|
metal response element binding transcription factor 2 |
| chr20_+_48921701 | 0.15 |
ENST00000371917.5
|
ARFGEF2
|
ADP ribosylation factor guanine nucleotide exchange factor 2 |
| chr13_+_48303709 | 0.15 |
ENST00000646097.1
ENST00000650461.1 ENST00000267163.6 |
RB1
|
RB transcriptional corepressor 1 |
| chr2_-_99489955 | 0.15 |
ENST00000393445.7
ENST00000258428.8 |
REV1
|
REV1 DNA directed polymerase |
| chr13_+_114281577 | 0.15 |
ENST00000375299.8
ENST00000492270.1 ENST00000351487.5 |
UPF3A
|
UPF3A regulator of nonsense mediated mRNA decay |
| chr16_-_58294976 | 0.15 |
ENST00000543437.5
ENST00000569079.1 |
PRSS54
|
serine protease 54 |
| chr11_+_1834804 | 0.15 |
ENST00000341958.3
|
SYT8
|
synaptotagmin 8 |
| chr2_-_191151568 | 0.15 |
ENST00000358470.8
ENST00000432798.1 ENST00000450994.1 |
STAT4
|
signal transducer and activator of transcription 4 |
| chr22_+_22734577 | 0.15 |
ENST00000390310.3
|
IGLV2-18
|
immunoglobulin lambda variable 2-18 |
| chr15_+_86079863 | 0.15 |
ENST00000614907.3
ENST00000441037.7 |
AGBL1
|
ATP/GTP binding protein like 1 |
| chr6_-_31777319 | 0.14 |
ENST00000375688.5
|
VWA7
|
von Willebrand factor A domain containing 7 |
| chr9_+_88535101 | 0.14 |
ENST00000618633.1
ENST00000375854.7 ENST00000375855.3 |
NXNL2
|
nucleoredoxin like 2 |
| chr19_-_40690553 | 0.14 |
ENST00000598779.5
|
NUMBL
|
NUMB like endocytic adaptor protein |
| chr22_+_22409755 | 0.14 |
ENST00000390299.2
|
IGLV1-40
|
immunoglobulin lambda variable 1-40 |
| chr1_+_241652275 | 0.14 |
ENST00000366552.6
ENST00000437684.7 |
WDR64
|
WD repeat domain 64 |
| chr1_+_39081316 | 0.14 |
ENST00000484793.5
|
MACF1
|
microtubule actin crosslinking factor 1 |
| chr19_+_40611863 | 0.14 |
ENST00000601032.5
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr9_-_86282061 | 0.14 |
ENST00000311534.6
|
ISCA1
|
iron-sulfur cluster assembly 1 |
| chr19_-_45322867 | 0.14 |
ENST00000221476.4
|
CKM
|
creatine kinase, M-type |
| chr7_-_56092932 | 0.14 |
ENST00000446428.5
ENST00000432123.5 ENST00000297373.7 |
PHKG1
|
phosphorylase kinase catalytic subunit gamma 1 |
| chr7_-_56092974 | 0.13 |
ENST00000452681.6
ENST00000537360.5 |
PHKG1
|
phosphorylase kinase catalytic subunit gamma 1 |
| chr11_+_1834415 | 0.13 |
ENST00000381968.7
ENST00000381978.7 |
SYT8
|
synaptotagmin 8 |
| chr9_+_27109135 | 0.13 |
ENST00000519097.5
ENST00000615002.4 |
TEK
|
TEK receptor tyrosine kinase |
| chr3_-_183555696 | 0.13 |
ENST00000341319.8
|
KLHL6
|
kelch like family member 6 |
| chr6_-_127519191 | 0.13 |
ENST00000525778.5
|
SOGA3
|
SOGA family member 3 |
| chr9_-_86282511 | 0.13 |
ENST00000375991.9
ENST00000326094.4 |
ISCA1
|
iron-sulfur cluster assembly 1 |
| chr12_+_10212867 | 0.13 |
ENST00000545047.5
ENST00000266458.10 ENST00000629504.1 ENST00000543602.5 ENST00000545887.1 |
GABARAPL1
|
GABA type A receptor associated protein like 1 |
| chr15_+_78266181 | 0.13 |
ENST00000446172.2
|
DNAJA4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr17_-_46192767 | 0.13 |
ENST00000262419.10
|
KANSL1
|
KAT8 regulatory NSL complex subunit 1 |
| chr22_-_37188233 | 0.13 |
ENST00000434784.1
ENST00000337843.7 |
C1QTNF6
|
C1q and TNF related 6 |
| chr7_-_73738831 | 0.13 |
ENST00000395147.9
ENST00000437775.7 |
ABHD11
|
abhydrolase domain containing 11 |
| chr4_+_7192519 | 0.13 |
ENST00000507866.6
|
SORCS2
|
sortilin related VPS10 domain containing receptor 2 |
| chr17_-_60392113 | 0.13 |
ENST00000300896.9
ENST00000589335.5 |
USP32
|
ubiquitin specific peptidase 32 |
| chr8_-_130016395 | 0.13 |
ENST00000523509.5
|
CYRIB
|
CYFIP related Rac1 interactor B |
| chr3_+_12289061 | 0.13 |
ENST00000652522.1
ENST00000652431.1 ENST00000652098.1 ENST00000651735.1 ENST00000397026.7 |
PPARG
|
peroxisome proliferator activated receptor gamma |
| chr2_-_229921963 | 0.13 |
ENST00000343290.5
ENST00000389044.8 ENST00000675453.1 ENST00000675903.1 ENST00000283943.9 |
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr15_-_34337772 | 0.13 |
ENST00000354181.8
|
SLC12A6
|
solute carrier family 12 member 6 |
| chr1_+_93079264 | 0.13 |
ENST00000370298.9
ENST00000370303.4 |
MTF2
|
metal response element binding transcription factor 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0031550 | positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) |
| 0.3 | 0.8 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.2 | 0.5 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.2 | 0.5 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.2 | 1.4 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.1 | 0.6 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 1.0 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 1.4 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 1.0 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 1.0 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.4 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.1 | 0.7 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.1 | 0.5 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.1 | 0.3 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.1 | 0.4 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.1 | 0.8 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.6 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.2 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.1 | 0.2 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.3 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.3 | GO:1904045 | cellular response to aldosterone(GO:1904045) |
| 0.1 | 0.7 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.1 | 0.3 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.1 | 0.3 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.1 | 0.3 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 0.3 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.3 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.2 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.1 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.3 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.0 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.1 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.0 | 0.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.2 | GO:0019477 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 0.5 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.9 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.3 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 0.3 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.2 | GO:0061052 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.0 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 0.1 | GO:0035822 | release from viral latency(GO:0019046) gene conversion(GO:0035822) regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.2 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.1 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.4 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) fourth ventricle development(GO:0021592) |
| 0.0 | 0.1 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.4 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.4 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.0 | 0.1 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:1900114 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) positive regulation of histone H3-K9 trimethylation(GO:1900114) |
| 0.0 | 0.7 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.1 | GO:1904719 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.6 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.4 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.1 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.1 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.2 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.5 | GO:2000352 | negative regulation of endothelial cell apoptotic process(GO:2000352) |
| 0.0 | 0.2 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.2 | GO:0097240 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.4 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 2.0 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.3 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.0 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 0.1 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.1 | GO:0006203 | dGTP catabolic process(GO:0006203) dATP catabolic process(GO:0046061) |
| 0.0 | 0.2 | GO:1903286 | regulation of potassium ion import(GO:1903286) |
| 0.0 | 0.0 | GO:0046671 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.1 | GO:0060083 | cellular potassium ion homeostasis(GO:0030007) smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.2 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.1 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.0 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.7 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 1.3 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0071752 | dimeric IgA immunoglobulin complex(GO:0071750) secretory dimeric IgA immunoglobulin complex(GO:0071752) |
| 0.2 | 0.8 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.3 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.1 | 0.4 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.2 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.1 | 0.4 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.3 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 1.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.2 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.3 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.1 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 1.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.6 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:0001520 | outer dense fiber(GO:0001520) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.2 | 1.4 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.2 | 0.8 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.9 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.4 | GO:0052857 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.1 | 0.4 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.1 | 0.4 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.1 | 0.8 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.1 | 0.4 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.1 | 0.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 1.0 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.1 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.1 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.2 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.0 | 0.3 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.3 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0047693 | GTP diphosphatase activity(GO:0036219) 2-hydroxy-adenosine triphosphate pyrophosphatase activity(GO:0044713) 2-hydroxy-(deoxy)adenosine-triphosphate pyrophosphatase activity(GO:0044714) ATP diphosphatase activity(GO:0047693) |
| 0.0 | 0.5 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.0 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 1.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.7 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.1 | GO:0051538 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.1 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 1.1 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.0 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.3 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 1.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.8 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 1.0 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.5 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |