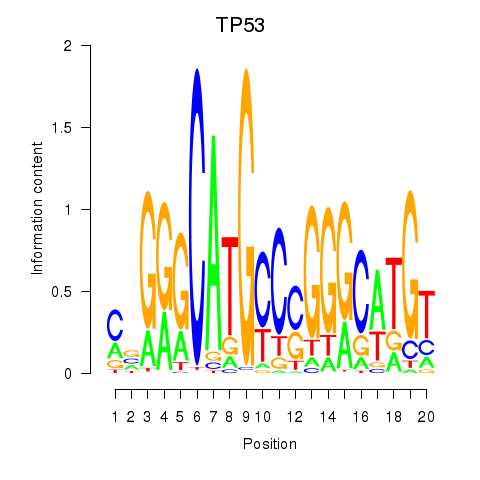
|
chr5_-_132490750
Show fit
|
1.13 |
ENST00000437654.6
ENST00000245414.9
ENST00000680139.1
ENST00000680352.1
ENST00000679440.1
ENST00000680903.1
|
IRF1
|
interferon regulatory factor 1
|
|
chr6_+_36676489
Show fit
|
1.05 |
ENST00000448526.6
|
CDKN1A
|
cyclin dependent kinase inhibitor 1A
|
|
chr6_+_36676455
Show fit
|
1.05 |
ENST00000615513.4
|
CDKN1A
|
cyclin dependent kinase inhibitor 1A
|
|
chr16_+_57372481
Show fit
|
0.86 |
ENST00000006053.7
|
CX3CL1
|
C-X3-C motif chemokine ligand 1
|
|
chr16_+_57372465
Show fit
|
0.85 |
ENST00000563383.1
|
CX3CL1
|
C-X3-C motif chemokine ligand 1
|
|
chr12_+_22625357
Show fit
|
0.71 |
ENST00000545979.2
|
ETNK1
|
ethanolamine kinase 1
|
|
chr8_-_23164020
Show fit
|
0.64 |
ENST00000312584.4
|
TNFRSF10D
|
TNF receptor superfamily member 10d
|
|
chr18_+_59899988
Show fit
|
0.58 |
ENST00000316660.7
ENST00000269518.9
|
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1
|
|
chr19_+_507487
Show fit
|
0.57 |
ENST00000359315.6
|
TPGS1
|
tubulin polyglutamylase complex subunit 1
|
|
chr8_-_23069012
Show fit
|
0.56 |
ENST00000347739.3
ENST00000276431.9
|
TNFRSF10B
|
TNF receptor superfamily member 10b
|
|
chr6_-_30742203
Show fit
|
0.53 |
ENST00000416018.5
ENST00000445853.5
ENST00000413165.5
ENST00000418160.5
|
FLOT1
|
flotillin 1
|
|
chr5_+_10564064
Show fit
|
0.49 |
ENST00000296657.7
|
ANKRD33B
|
ankyrin repeat domain 33B
|
|
chr10_+_129467178
Show fit
|
0.48 |
ENST00000306010.8
ENST00000651593.1
|
MGMT
|
O-6-methylguanine-DNA methyltransferase
|
|
chr19_-_47232649
Show fit
|
0.47 |
ENST00000449228.5
ENST00000300880.11
ENST00000341983.8
|
BBC3
|
BCL2 binding component 3
|
|
chr2_-_234497035
Show fit
|
0.46 |
ENST00000390645.2
ENST00000339728.6
|
ARL4C
|
ADP ribosylation factor like GTPase 4C
|
|
chr16_-_4614876
Show fit
|
0.46 |
ENST00000591401.5
ENST00000283474.12
ENST00000591897.5
|
UBALD1
|
UBA like domain containing 1
|
|
chr5_+_122311740
Show fit
|
0.44 |
ENST00000506272.5
ENST00000508681.5
ENST00000509154.6
|
SNCAIP
|
synuclein alpha interacting protein
|
|
chr8_-_10730498
Show fit
|
0.44 |
ENST00000304501.2
|
SOX7
|
SRY-box transcription factor 7
|
|
chr8_+_23102913
Show fit
|
0.41 |
ENST00000356864.4
|
TNFRSF10C
|
TNF receptor superfamily member 10c
|
|
chr16_+_57619942
Show fit
|
0.41 |
ENST00000568908.5
ENST00000568909.5
ENST00000566778.5
ENST00000561988.5
|
ADGRG1
|
adhesion G protein-coupled receptor G1
|
|
chr5_+_122312229
Show fit
|
0.41 |
ENST00000261368.13
|
SNCAIP
|
synuclein alpha interacting protein
|
|
chr5_+_122312164
Show fit
|
0.40 |
ENST00000514497.6
ENST00000261367.11
|
SNCAIP
|
synuclein alpha interacting protein
|
|
chr16_+_57620077
Show fit
|
0.40 |
ENST00000567835.5
ENST00000569372.5
ENST00000563548.5
ENST00000562003.5
|
ADGRG1
|
adhesion G protein-coupled receptor G1
|
|
chr15_+_88621290
Show fit
|
0.37 |
ENST00000332810.4
ENST00000559528.1
|
AEN
|
apoptosis enhancing nuclease
|
|
chr7_+_154305256
Show fit
|
0.37 |
ENST00000619756.4
|
DPP6
|
dipeptidyl peptidase like 6
|
|
chr1_-_17011891
Show fit
|
0.37 |
ENST00000341676.9
ENST00000326735.13
ENST00000452699.5
|
ATP13A2
|
ATPase cation transporting 13A2
|
|
chr17_+_78214186
Show fit
|
0.35 |
ENST00000301633.8
ENST00000350051.8
ENST00000374948.6
ENST00000590449.1
|
BIRC5
|
baculoviral IAP repeat containing 5
|
|
chr7_+_48088596
Show fit
|
0.34 |
ENST00000416681.5
ENST00000331803.8
ENST00000432131.5
|
UPP1
|
uridine phosphorylase 1
|
|
chr20_+_35172046
Show fit
|
0.34 |
ENST00000216968.5
|
PROCR
|
protein C receptor
|
|
chr9_+_136952256
Show fit
|
0.34 |
ENST00000371633.8
|
LCN12
|
lipocalin 12
|
|
chr20_-_5119945
Show fit
|
0.34 |
ENST00000379143.10
|
PCNA
|
proliferating cell nuclear antigen
|
|
chr8_+_42338454
Show fit
|
0.34 |
ENST00000532157.5
ENST00000520008.5
|
POLB
|
DNA polymerase beta
|
|
chrX_+_152917830
Show fit
|
0.33 |
ENST00000318529.12
|
ZNF185
|
zinc finger protein 185 with LIM domain
|
|
chr11_-_44950839
Show fit
|
0.33 |
ENST00000395648.7
ENST00000531928.6
|
TP53I11
|
tumor protein p53 inducible protein 11
|
|
chr9_+_136952896
Show fit
|
0.32 |
ENST00000371632.7
|
LCN12
|
lipocalin 12
|
|
chr3_+_32106612
Show fit
|
0.31 |
ENST00000282541.10
ENST00000425459.5
ENST00000431009.1
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1 like
|
|
chr17_+_76376581
Show fit
|
0.30 |
ENST00000591651.5
ENST00000545180.5
|
SPHK1
|
sphingosine kinase 1
|
|
chr1_-_54406385
Show fit
|
0.30 |
ENST00000610401.5
|
SSBP3
|
single stranded DNA binding protein 3
|
|
chr2_-_288056
Show fit
|
0.30 |
ENST00000403610.9
|
ALKAL2
|
ALK and LTK ligand 2
|
|
chr20_+_44355692
Show fit
|
0.29 |
ENST00000316673.8
ENST00000609795.5
ENST00000457232.5
ENST00000609262.5
|
HNF4A
|
hepatocyte nuclear factor 4 alpha
|
|
chr12_-_6342020
Show fit
|
0.29 |
ENST00000540022.5
ENST00000536194.1
|
TNFRSF1A
|
TNF receptor superfamily member 1A
|
|
chr9_-_133121148
Show fit
|
0.28 |
ENST00000372047.7
|
RALGDS
|
ral guanine nucleotide dissociation stimulator
|
|
chr4_-_10116324
Show fit
|
0.27 |
ENST00000508079.1
|
WDR1
|
WD repeat domain 1
|
|
chr1_-_156741067
Show fit
|
0.27 |
ENST00000361531.6
ENST00000412846.5
|
MRPL24
|
mitochondrial ribosomal protein L24
|
|
chr19_+_35371290
Show fit
|
0.27 |
ENST00000597214.1
|
GPR42
|
G protein-coupled receptor 42
|
|
chr11_-_73598183
Show fit
|
0.26 |
ENST00000064778.8
|
FAM168A
|
family with sequence similarity 168 member A
|
|
chr17_+_78214286
Show fit
|
0.26 |
ENST00000592734.5
ENST00000587746.5
|
BIRC5
|
baculoviral IAP repeat containing 5
|
|
chr1_+_156082563
Show fit
|
0.26 |
ENST00000368301.6
|
LMNA
|
lamin A/C
|
|
chr10_+_88990736
Show fit
|
0.25 |
ENST00000357339.6
ENST00000652046.1
ENST00000355279.2
|
FAS
|
Fas cell surface death receptor
|
|
chr19_+_48954850
Show fit
|
0.25 |
ENST00000345358.12
ENST00000539787.2
ENST00000415969.6
ENST00000354470.7
ENST00000506183.5
ENST00000391871.4
ENST00000293288.12
|
BAX
|
BCL2 associated X, apoptosis regulator
|
|
chr1_-_156741124
Show fit
|
0.25 |
ENST00000368211.8
|
MRPL24
|
mitochondrial ribosomal protein L24
|
|
chr11_+_65111845
Show fit
|
0.25 |
ENST00000526809.5
ENST00000524986.5
ENST00000534371.5
ENST00000279263.14
ENST00000525385.5
ENST00000345348.9
ENST00000531321.5
ENST00000529414.5
ENST00000526085.5
ENST00000530750.5
|
TM7SF2
|
transmembrane 7 superfamily member 2
|
|
chr19_-_42242526
Show fit
|
0.25 |
ENST00000222330.8
ENST00000676537.1
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr6_-_30744537
Show fit
|
0.25 |
ENST00000259874.6
ENST00000376377.2
|
IER3
|
immediate early response 3
|
|
chr7_+_101127095
Show fit
|
0.25 |
ENST00000223095.5
|
SERPINE1
|
serpin family E member 1
|
|
chr9_-_133121228
Show fit
|
0.25 |
ENST00000372050.8
|
RALGDS
|
ral guanine nucleotide dissociation stimulator
|
|
chr11_-_73598067
Show fit
|
0.23 |
ENST00000450446.6
ENST00000356467.5
|
FAM168A
|
family with sequence similarity 168 member A
|
|
chr9_-_135961310
Show fit
|
0.23 |
ENST00000371756.4
|
UBAC1
|
UBA domain containing 1
|
|
chr1_-_53945567
Show fit
|
0.22 |
ENST00000371378.6
|
HSPB11
|
heat shock protein family B (small) member 11
|
|
chr10_-_125158704
Show fit
|
0.22 |
ENST00000531469.5
|
CTBP2
|
C-terminal binding protein 2
|
|
chr11_-_75306688
Show fit
|
0.22 |
ENST00000532525.1
|
ARRB1
|
arrestin beta 1
|
|
chr1_-_53945584
Show fit
|
0.22 |
ENST00000371377.3
|
HSPB11
|
heat shock protein family B (small) member 11
|
|
chr17_+_20579724
Show fit
|
0.22 |
ENST00000661883.1
ENST00000399044.1
|
CDRT15L2
|
CMT1A duplicated region transcript 15 like 2
|
|
chr11_-_73982830
Show fit
|
0.22 |
ENST00000536983.5
ENST00000663595.2
ENST00000310473.9
|
UCP2
|
uncoupling protein 2
|
|
chr20_-_23826721
Show fit
|
0.21 |
ENST00000304725.3
|
CST2
|
cystatin SA
|
|
chr12_-_54648356
Show fit
|
0.20 |
ENST00000293371.11
ENST00000456047.2
|
DCD
|
dermcidin
|
|
chr16_+_77722502
Show fit
|
0.20 |
ENST00000564085.5
ENST00000268533.9
ENST00000568787.5
ENST00000437314.3
ENST00000563839.1
|
NUDT7
|
nudix hydrolase 7
|
|
chr16_+_2029636
Show fit
|
0.19 |
ENST00000561844.5
|
SLC9A3R2
|
SLC9A3 regulator 2
|
|
chr22_+_19718390
Show fit
|
0.19 |
ENST00000383045.7
ENST00000438754.6
|
SEPTIN5
|
septin 5
|
|
chr2_-_25982471
Show fit
|
0.19 |
ENST00000264712.8
|
KIF3C
|
kinesin family member 3C
|
|
chr16_-_67966793
Show fit
|
0.18 |
ENST00000541864.6
|
SLC12A4
|
solute carrier family 12 member 4
|
|
chr19_-_8698705
Show fit
|
0.18 |
ENST00000612068.1
|
ACTL9
|
actin like 9
|
|
chr1_-_54053394
Show fit
|
0.18 |
ENST00000452421.5
ENST00000420738.5
ENST00000440019.5
|
TMEM59
|
transmembrane protein 59
|
|
chr16_+_77722473
Show fit
|
0.18 |
ENST00000613075.4
|
NUDT7
|
nudix hydrolase 7
|
|
chr3_-_42875871
Show fit
|
0.18 |
ENST00000316161.6
ENST00000437102.1
|
CYP8B1
|
cytochrome P450 family 8 subfamily B member 1
|
|
chr14_+_50533026
Show fit
|
0.17 |
ENST00000441560.6
|
ATL1
|
atlastin GTPase 1
|
|
chr20_+_4148779
Show fit
|
0.17 |
ENST00000621355.4
ENST00000339123.10
ENST00000278795.7
ENST00000305958.9
ENST00000346595.6
|
SMOX
|
spermine oxidase
|
|
chr1_+_202416826
Show fit
|
0.17 |
ENST00000466968.1
|
PPP1R12B
|
protein phosphatase 1 regulatory subunit 12B
|
|
chr4_+_6269869
Show fit
|
0.17 |
ENST00000506362.2
|
WFS1
|
wolframin ER transmembrane glycoprotein
|
|
chr7_+_73328152
Show fit
|
0.17 |
ENST00000252037.5
ENST00000431982.6
|
FKBP6
|
FKBP prolyl isomerase family member 6 (inactive)
|
|
chr1_-_201469151
Show fit
|
0.17 |
ENST00000367311.5
ENST00000367309.1
|
PHLDA3
|
pleckstrin homology like domain family A member 3
|
|
chr7_-_100168602
Show fit
|
0.16 |
ENST00000423751.2
ENST00000360039.9
|
GAL3ST4
|
galactose-3-O-sulfotransferase 4
|
|
chr7_+_73328177
Show fit
|
0.16 |
ENST00000442793.5
ENST00000413573.6
|
FKBP6
|
FKBP prolyl isomerase family member 6 (inactive)
|
|
chr1_-_1307861
Show fit
|
0.16 |
ENST00000354700.10
|
ACAP3
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3
|
|
chr19_-_5791155
Show fit
|
0.16 |
ENST00000309061.12
|
DUS3L
|
dihydrouridine synthase 3 like
|
|
chr5_-_95961830
Show fit
|
0.16 |
ENST00000513343.1
ENST00000237853.9
|
ELL2
|
elongation factor for RNA polymerase II 2
|
|
chr16_+_142725
Show fit
|
0.15 |
ENST00000652335.1
|
HBZ
|
hemoglobin subunit zeta
|
|
chr1_-_6393339
Show fit
|
0.15 |
ENST00000608083.5
|
ACOT7
|
acyl-CoA thioesterase 7
|
|
chr6_-_132714045
Show fit
|
0.15 |
ENST00000367928.5
|
VNN1
|
vanin 1
|
|
chr8_-_142777174
Show fit
|
0.15 |
ENST00000652477.1
ENST00000614491.1
ENST00000613110.4
|
LYNX1
|
Ly6/neurotoxin 1
|
|
chr14_-_106235582
Show fit
|
0.15 |
ENST00000390607.2
|
IGHV3-21
|
immunoglobulin heavy variable 3-21
|
|
chr20_-_23637947
Show fit
|
0.15 |
ENST00000376925.8
|
CST3
|
cystatin C
|
|
chr14_-_103522696
Show fit
|
0.15 |
ENST00000553878.5
ENST00000348956.7
ENST00000557530.1
|
CKB
|
creatine kinase B
|
|
chr2_-_241508630
Show fit
|
0.15 |
ENST00000403346.7
ENST00000535007.5
|
STK25
|
serine/threonine kinase 25
|
|
chr11_+_61950063
Show fit
|
0.15 |
ENST00000534553.5
|
BEST1
|
bestrophin 1
|
|
chr7_-_44885446
Show fit
|
0.15 |
ENST00000395699.5
|
PURB
|
purine rich element binding protein B
|
|
chr19_-_55179390
Show fit
|
0.15 |
ENST00000590851.5
|
SYT5
|
synaptotagmin 5
|
|
chr10_+_132186937
Show fit
|
0.14 |
ENST00000338492.9
|
DPYSL4
|
dihydropyrimidinase like 4
|
|
chr12_-_6342066
Show fit
|
0.14 |
ENST00000162749.7
ENST00000440083.6
|
TNFRSF1A
|
TNF receptor superfamily member 1A
|
|
chr1_-_54053461
Show fit
|
0.14 |
ENST00000371341.5
|
TMEM59
|
transmembrane protein 59
|
|
chr3_-_39281663
Show fit
|
0.14 |
ENST00000358309.3
|
CX3CR1
|
C-X3-C motif chemokine receptor 1
|
|
chr11_-_65859282
Show fit
|
0.14 |
ENST00000526975.1
ENST00000531413.5
ENST00000525451.6
|
CFL1
|
cofilin 1
|
|
chr15_-_42094560
Show fit
|
0.14 |
ENST00000290472.4
|
PLA2G4D
|
phospholipase A2 group IVD
|
|
chr20_+_58692767
Show fit
|
0.13 |
ENST00000356091.11
|
NPEPL1
|
aminopeptidase like 1
|
|
chr19_+_55488404
Show fit
|
0.13 |
ENST00000594321.5
ENST00000389623.11
|
SSC5D
|
scavenger receptor cysteine rich family member with 5 domains
|
|
chr18_-_31042808
Show fit
|
0.13 |
ENST00000434452.5
|
DSC3
|
desmocollin 3
|
|
chr2_+_168456215
Show fit
|
0.13 |
ENST00000392687.4
ENST00000305747.11
|
CERS6
|
ceramide synthase 6
|
|
chr10_+_90871946
Show fit
|
0.13 |
ENST00000413330.5
ENST00000371703.8
ENST00000277882.7
|
RPP30
|
ribonuclease P/MRP subunit p30
|
|
chr1_-_160862880
Show fit
|
0.13 |
ENST00000368034.9
|
CD244
|
CD244 molecule
|
|
chr12_+_12813316
Show fit
|
0.13 |
ENST00000352940.8
ENST00000358007.7
ENST00000544400.1
|
DDX47
|
DEAD-box helicase 47
|
|
chrX_+_71254781
Show fit
|
0.12 |
ENST00000677446.1
|
NONO
|
non-POU domain containing octamer binding
|
|
chr11_-_47378527
Show fit
|
0.12 |
ENST00000378538.8
|
SPI1
|
Spi-1 proto-oncogene
|
|
chr1_+_107139996
Show fit
|
0.12 |
ENST00000370073.6
ENST00000370074.8
|
NTNG1
|
netrin G1
|
|
chr2_-_241617464
Show fit
|
0.12 |
ENST00000402545.5
ENST00000402136.5
|
THAP4
|
THAP domain containing 4
|
|
chr9_-_127122623
Show fit
|
0.12 |
ENST00000373417.1
ENST00000373425.8
|
ANGPTL2
|
angiopoietin like 2
|
|
chr1_-_45339995
Show fit
|
0.12 |
ENST00000488731.6
ENST00000435155.1
|
MUTYH
|
mutY DNA glycosylase
|
|
chr1_-_45340381
Show fit
|
0.12 |
ENST00000412971.5
ENST00000372110.7
ENST00000372098.7
ENST00000672818.3
ENST00000529984.5
ENST00000372115.7
|
MUTYH
|
mutY DNA glycosylase
|
|
chr2_-_241508568
Show fit
|
0.11 |
ENST00000426941.1
ENST00000316586.9
ENST00000405585.5
ENST00000420551.1
ENST00000429279.5
ENST00000442307.5
|
STK25
|
serine/threonine kinase 25
|
|
chr3_+_44338452
Show fit
|
0.11 |
ENST00000396078.7
|
TCAIM
|
T cell activation inhibitor, mitochondrial
|
|
chr10_+_11742361
Show fit
|
0.11 |
ENST00000379215.9
ENST00000420401.5
|
ECHDC3
|
enoyl-CoA hydratase domain containing 3
|
|
chr1_-_3611470
Show fit
|
0.11 |
ENST00000356575.9
|
MEGF6
|
multiple EGF like domains 6
|
|
chr4_-_55125585
Show fit
|
0.11 |
ENST00000263923.5
|
KDR
|
kinase insert domain receptor
|
|
chr20_+_49812697
Show fit
|
0.11 |
ENST00000417961.5
|
SLC9A8
|
solute carrier family 9 member A8
|
|
chr22_+_46674593
Show fit
|
0.11 |
ENST00000408031.1
|
GRAMD4
|
GRAM domain containing 4
|
|
chr4_+_677922
Show fit
|
0.11 |
ENST00000400159.6
|
MYL5
|
myosin light chain 5
|
|
chr1_+_113905156
Show fit
|
0.11 |
ENST00000650596.1
|
DCLRE1B
|
DNA cross-link repair 1B
|
|
chr20_+_49812818
Show fit
|
0.11 |
ENST00000361573.3
|
SLC9A8
|
solute carrier family 9 member A8
|
|
chr5_-_176610104
Show fit
|
0.11 |
ENST00000303991.5
|
GPRIN1
|
G protein regulated inducer of neurite outgrowth 1
|
|
chr15_-_63156774
Show fit
|
0.11 |
ENST00000462430.5
|
RPS27L
|
ribosomal protein S27 like
|
|
chr2_-_241508547
Show fit
|
0.11 |
ENST00000405883.7
|
STK25
|
serine/threonine kinase 25
|
|
chr20_+_32358303
Show fit
|
0.11 |
ENST00000651418.1
ENST00000375687.10
ENST00000542461.5
ENST00000613218.4
ENST00000646367.1
ENST00000620121.4
|
ASXL1
|
ASXL transcriptional regulator 1
|
|
chr19_-_42217667
Show fit
|
0.11 |
ENST00000336034.8
ENST00000596251.6
ENST00000598200.1
ENST00000598727.5
|
DEDD2
|
death effector domain containing 2
|
|
chr20_-_1393074
Show fit
|
0.11 |
ENST00000614856.2
ENST00000678408.1
ENST00000618612.5
ENST00000439640.5
ENST00000381719.8
ENST00000677533.1
|
FKBP1A
|
FKBP prolyl isomerase 1A
|
|
chr8_-_23225061
Show fit
|
0.11 |
ENST00000613472.1
ENST00000221132.8
|
TNFRSF10A
|
TNF receptor superfamily member 10a
|
|
chr8_-_12194067
Show fit
|
0.10 |
ENST00000524571.6
ENST00000533852.6
ENST00000533513.1
ENST00000448228.6
ENST00000534520.5
|
FAM86B1
|
family with sequence similarity 86 member B1
|
|
chr3_+_44338119
Show fit
|
0.10 |
ENST00000383746.7
ENST00000417237.5
|
TCAIM
|
T cell activation inhibitor, mitochondrial
|
|
chr1_-_2526585
Show fit
|
0.10 |
ENST00000378466.9
ENST00000435556.8
|
PANK4
|
pantothenate kinase 4 (inactive)
|
|
chr11_-_3057386
Show fit
|
0.10 |
ENST00000529772.5
ENST00000278224.13
ENST00000380525.9
|
CARS1
|
cysteinyl-tRNA synthetase 1
|
|
chr14_-_74612226
Show fit
|
0.10 |
ENST00000261978.9
|
LTBP2
|
latent transforming growth factor beta binding protein 2
|
|
chr1_-_45340080
Show fit
|
0.10 |
ENST00000354383.10
ENST00000355498.6
ENST00000531105.5
|
MUTYH
|
mutY DNA glycosylase
|
|
chr11_-_47378391
Show fit
|
0.10 |
ENST00000227163.8
|
SPI1
|
Spi-1 proto-oncogene
|
|
chr11_-_65540610
Show fit
|
0.10 |
ENST00000532661.5
|
LTBP3
|
latent transforming growth factor beta binding protein 3
|
|
chr16_+_2496032
Show fit
|
0.10 |
ENST00000564543.1
|
ENSG00000260272.1
|
novel protein
|
|
chr4_+_51843063
Show fit
|
0.10 |
ENST00000381441.7
ENST00000334635.10
|
DCUN1D4
|
defective in cullin neddylation 1 domain containing 4
|
|
chr19_-_14117729
Show fit
|
0.10 |
ENST00000590853.5
ENST00000308677.9
|
PRKACA
|
protein kinase cAMP-activated catalytic subunit alpha
|
|
chr5_-_113488415
Show fit
|
0.09 |
ENST00000408903.7
|
MCC
|
MCC regulator of WNT signaling pathway
|
|
chr3_+_184176778
Show fit
|
0.09 |
ENST00000439647.5
|
AP2M1
|
adaptor related protein complex 2 subunit mu 1
|
|
chr17_-_1516621
Show fit
|
0.09 |
ENST00000574561.1
|
INPP5K
|
inositol polyphosphate-5-phosphatase K
|
|
chr19_-_51646800
Show fit
|
0.09 |
ENST00000599649.5
ENST00000429354.3
ENST00000360844.6
|
SIGLEC5
SIGLEC14
|
sialic acid binding Ig like lectin 5
sialic acid binding Ig like lectin 14
|
|
chr5_+_163437569
Show fit
|
0.09 |
ENST00000512163.5
ENST00000393929.5
ENST00000510097.5
ENST00000340828.7
ENST00000511490.4
ENST00000510664.5
|
CCNG1
|
cyclin G1
|
|
chr17_-_47841173
Show fit
|
0.09 |
ENST00000584123.5
ENST00000578323.1
ENST00000290216.14
ENST00000407215.7
|
SCRN2
|
secernin 2
|
|
chr16_-_31094727
Show fit
|
0.09 |
ENST00000300851.10
ENST00000394975.3
|
VKORC1
|
vitamin K epoxide reductase complex subunit 1
|
|
chr9_+_36136416
Show fit
|
0.09 |
ENST00000396613.7
|
GLIPR2
|
GLI pathogenesis related 2
|
|
chr20_-_34955635
Show fit
|
0.09 |
ENST00000644793.1
ENST00000642498.1
ENST00000646735.1
ENST00000644608.1
ENST00000651619.1
|
GSS
|
glutathione synthetase
|
|
chr16_-_31094890
Show fit
|
0.09 |
ENST00000532364.1
ENST00000529564.1
ENST00000319788.11
ENST00000354895.4
|
ENSG00000255439.6
VKORC1
|
novel protein, VKORC1 and PRSS53 readthrough
vitamin K epoxide reductase complex subunit 1
|
|
chr5_+_177134159
Show fit
|
0.09 |
ENST00000511258.5
ENST00000347982.8
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chr11_+_65879791
Show fit
|
0.09 |
ENST00000528419.6
ENST00000307886.8
ENST00000526034.2
ENST00000679584.1
ENST00000680443.1
ENST00000680670.1
|
CTSW
|
cathepsin W
|
|
chr17_-_74859863
Show fit
|
0.09 |
ENST00000293190.10
|
GRIN2C
|
glutamate ionotropic receptor NMDA type subunit 2C
|
|
chr16_-_14694308
Show fit
|
0.09 |
ENST00000438167.8
|
PLA2G10
|
phospholipase A2 group X
|
|
chr12_-_57237090
Show fit
|
0.09 |
ENST00000556732.1
|
NDUFA4L2
|
NDUFA4 mitochondrial complex associated like 2
|
|
chr11_-_129192198
Show fit
|
0.08 |
ENST00000310343.13
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr1_-_209652329
Show fit
|
0.08 |
ENST00000367030.7
ENST00000356082.9
|
LAMB3
|
laminin subunit beta 3
|
|
chr18_-_36828771
Show fit
|
0.08 |
ENST00000589049.5
ENST00000587129.5
ENST00000590842.5
|
TPGS2
|
tubulin polyglutamylase complex subunit 2
|
|
chr16_-_31094549
Show fit
|
0.08 |
ENST00000394971.7
|
VKORC1
|
vitamin K epoxide reductase complex subunit 1
|
|
chr17_-_1516601
Show fit
|
0.08 |
ENST00000421807.7
ENST00000477910.5
ENST00000575172.5
|
INPP5K
|
inositol polyphosphate-5-phosphatase K
|
|
chr7_+_157139195
Show fit
|
0.08 |
ENST00000389103.4
|
UBE3C
|
ubiquitin protein ligase E3C
|
|
chr15_+_84817346
Show fit
|
0.08 |
ENST00000258888.6
|
ALPK3
|
alpha kinase 3
|
|
chr11_-_129192291
Show fit
|
0.08 |
ENST00000682385.1
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr17_-_1516699
Show fit
|
0.08 |
ENST00000320345.10
ENST00000406424.8
|
INPP5K
|
inositol polyphosphate-5-phosphatase K
|
|
chr1_+_54053573
Show fit
|
0.08 |
ENST00000234827.6
|
TCEANC2
|
transcription elongation factor A N-terminal and central domain containing 2
|
|
chr4_+_55346349
Show fit
|
0.08 |
ENST00000505210.1
|
SRD5A3
|
steroid 5 alpha-reductase 3
|
|
chr20_-_1393045
Show fit
|
0.08 |
ENST00000400137.9
ENST00000381715.4
|
FKBP1A
|
FKBP prolyl isomerase 1A
|
|
chr11_+_65181687
Show fit
|
0.08 |
ENST00000527739.5
ENST00000526966.5
ENST00000533129.5
ENST00000524773.5
|
CAPN1
|
calpain 1
|
|
chr18_-_36829154
Show fit
|
0.08 |
ENST00000614939.4
ENST00000610723.4
|
TPGS2
|
tubulin polyglutamylase complex subunit 2
|
|
chr4_+_185426234
Show fit
|
0.08 |
ENST00000511138.5
ENST00000511581.5
ENST00000378850.5
|
C4orf47
|
chromosome 4 open reading frame 47
|
|
chr22_-_36529136
Show fit
|
0.08 |
ENST00000216190.13
ENST00000455547.5
|
EIF3D
|
eukaryotic translation initiation factor 3 subunit D
|
|
chr1_-_12618198
Show fit
|
0.08 |
ENST00000616661.5
|
DHRS3
|
dehydrogenase/reductase 3
|
|
chr4_+_2536630
Show fit
|
0.08 |
ENST00000637812.2
|
FAM193A
|
family with sequence similarity 193 member A
|
|
chr14_+_24232921
Show fit
|
0.08 |
ENST00000557854.5
ENST00000399440.7
ENST00000559104.5
ENST00000456667.7
|
GMPR2
|
guanosine monophosphate reductase 2
|
|
chr22_-_20320009
Show fit
|
0.07 |
ENST00000405465.3
ENST00000248879.8
|
DGCR6L
|
DiGeorge syndrome critical region gene 6 like
|
|
chr3_+_184176949
Show fit
|
0.07 |
ENST00000432591.5
ENST00000431779.5
ENST00000621863.4
|
AP2M1
|
adaptor related protein complex 2 subunit mu 1
|
|
chr7_+_129434424
Show fit
|
0.07 |
ENST00000249344.7
ENST00000435494.2
|
STRIP2
|
striatin interacting protein 2
|
|
chr12_+_108562579
Show fit
|
0.07 |
ENST00000311893.14
ENST00000535729.5
ENST00000431221.6
ENST00000547005.5
ENST00000392807.8
ENST00000539593.1
|
ISCU
|
iron-sulfur cluster assembly enzyme
|
|
chr16_-_18433412
Show fit
|
0.07 |
ENST00000620440.4
|
NOMO2
|
NODAL modulator 2
|
|
chr11_-_2139382
Show fit
|
0.07 |
ENST00000416167.7
|
IGF2
|
insulin like growth factor 2
|
|
chr19_+_827823
Show fit
|
0.07 |
ENST00000233997.4
|
AZU1
|
azurocidin 1
|
|
chr1_-_156082412
Show fit
|
0.07 |
ENST00000532414.3
|
MEX3A
|
mex-3 RNA binding family member A
|
|
chr14_-_36521149
Show fit
|
0.07 |
ENST00000518149.5
|
NKX2-1
|
NK2 homeobox 1
|
|
chr22_-_36528897
Show fit
|
0.07 |
ENST00000405442.5
ENST00000402116.2
|
EIF3D
|
eukaryotic translation initiation factor 3 subunit D
|
|
chr7_-_38349972
Show fit
|
0.07 |
ENST00000390344.2
|
TRGV5
|
T cell receptor gamma variable 5
|
|
chr11_+_64306227
Show fit
|
0.06 |
ENST00000405666.5
ENST00000468670.2
|
ESRRA
|
estrogen related receptor alpha
|
|
chr22_-_22559073
Show fit
|
0.06 |
ENST00000420709.5
ENST00000398741.5
|
PRAME
|
PRAME nuclear receptor transcriptional regulator
|
|
chr10_-_71851313
Show fit
|
0.06 |
ENST00000394934.4
ENST00000610929.3
|
PSAP
|
prosaposin
|
|
chr2_-_241508318
Show fit
|
0.06 |
ENST00000439101.5
ENST00000424537.5
ENST00000401869.5
ENST00000436402.5
|
STK25
|
serine/threonine kinase 25
|
|
chr17_-_41525326
Show fit
|
0.06 |
ENST00000593096.1
|
KRT19
|
keratin 19
|
|
chr4_-_10116724
Show fit
|
0.06 |
ENST00000502702.5
|
WDR1
|
WD repeat domain 1
|
|
chr11_+_65181194
Show fit
|
0.06 |
ENST00000533820.5
|
CAPN1
|
calpain 1
|
|
chrX_-_66639022
Show fit
|
0.06 |
ENST00000374719.8
|
EDA2R
|
ectodysplasin A2 receptor
|
|
chr10_-_47484081
Show fit
|
0.06 |
ENST00000583448.2
ENST00000583874.5
ENST00000585281.6
|
ANXA8
|
annexin A8
|
|
chr1_-_248859067
Show fit
|
0.06 |
ENST00000496231.1
ENST00000306601.9
ENST00000366471.7
|
ZNF692
|
zinc finger protein 692
|