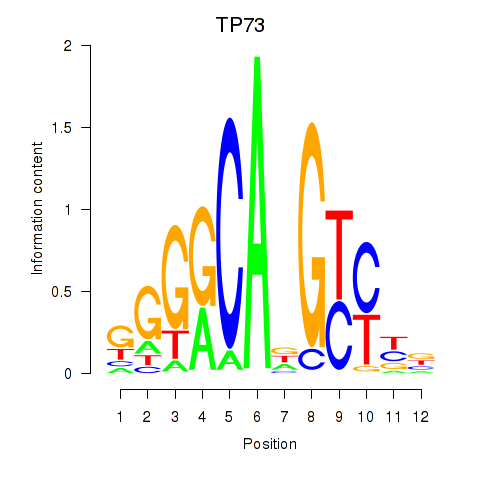
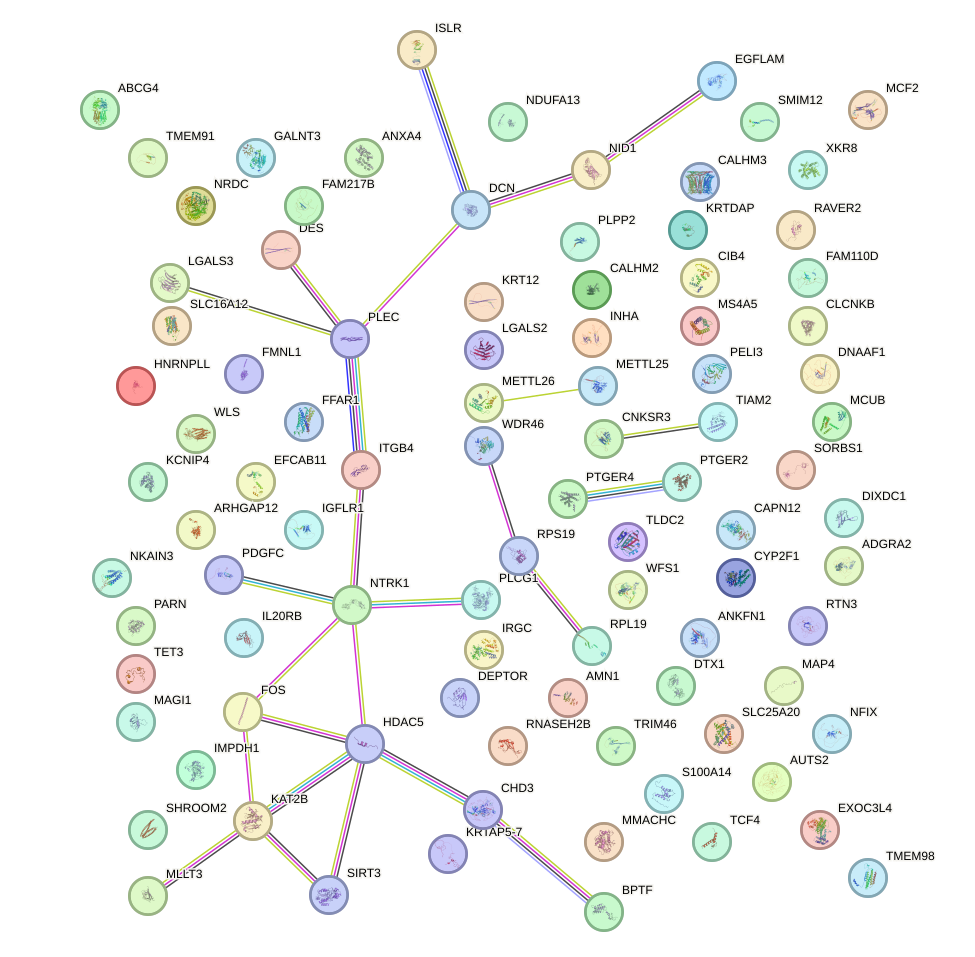
|
chr11_+_71527267
Show fit
|
0.95 |
ENST00000398536.6
|
KRTAP5-7
|
keratin associated protein 5-7
|
|
chr14_+_75279637
Show fit
|
0.61 |
ENST00000555686.1
ENST00000555672.1
|
FOS
|
Fos proto-oncogene, AP-1 transcription factor subunit
|
|
chr14_+_75278820
Show fit
|
0.57 |
ENST00000554617.1
ENST00000554212.5
ENST00000535987.5
ENST00000303562.9
ENST00000555242.1
|
FOS
|
Fos proto-oncogene, AP-1 transcription factor subunit
|
|
chr10_-_103452384
Show fit
|
0.51 |
ENST00000369788.7
|
CALHM2
|
calcium homeostasis modulator family member 2
|
|
chr10_-_103452356
Show fit
|
0.50 |
ENST00000260743.10
|
CALHM2
|
calcium homeostasis modulator family member 2
|
|
chrX_+_9912434
Show fit
|
0.42 |
ENST00000418909.6
|
SHROOM2
|
shroom family member 2
|
|
chr10_-_103479240
Show fit
|
0.41 |
ENST00000369783.4
|
CALHM3
|
calcium homeostasis modulator 3
|
|
chr3_+_20040437
Show fit
|
0.40 |
ENST00000263754.5
|
KAT2B
|
lysine acetyltransferase 2B
|
|
chr4_+_6269869
Show fit
|
0.39 |
ENST00000506362.2
|
WFS1
|
wolframin ER transmembrane glycoprotein
|
|
chr12_+_82358496
Show fit
|
0.33 |
ENST00000248306.8
ENST00000548200.5
|
METTL25
|
methyltransferase like 25
|
|
chr8_+_37796850
Show fit
|
0.31 |
ENST00000412232.3
|
ADGRA2
|
adhesion G protein-coupled receptor A2
|
|
chr8_+_37796906
Show fit
|
0.30 |
ENST00000315215.11
|
ADGRA2
|
adhesion G protein-coupled receptor A2
|
|
chr20_-_50636887
Show fit
|
0.29 |
ENST00000045083.6
|
RIPOR3
|
RIPOR family member 3
|
|
chr3_-_48898813
Show fit
|
0.28 |
ENST00000319017.5
ENST00000430379.5
|
SLC25A20
|
solute carrier family 25 member 20
|
|
chr19_-_291132
Show fit
|
0.27 |
ENST00000327790.7
|
PLPP2
|
phospholipid phosphatase 2
|
|
chr11_-_236821
Show fit
|
0.27 |
ENST00000529382.5
ENST00000528469.1
|
SIRT3
|
sirtuin 3
|
|
chr1_+_64745089
Show fit
|
0.26 |
ENST00000294428.7
ENST00000371072.8
|
RAVER2
|
ribonucleoprotein, PTB binding 2
|
|
chr3_-_66038537
Show fit
|
0.22 |
ENST00000483466.5
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1
|
|
chr20_+_59940362
Show fit
|
0.22 |
ENST00000360816.8
|
FAM217B
|
family with sequence similarity 217 member B
|
|
chr19_+_44777860
Show fit
|
0.22 |
ENST00000341505.4
ENST00000647358.2
|
CBLC
|
Cbl proto-oncogene C
|
|
chr8_+_62248591
Show fit
|
0.21 |
ENST00000519049.6
|
NKAIN3
|
sodium/potassium transporting ATPase interacting 3
|
|
chr2_-_165794190
Show fit
|
0.21 |
ENST00000392701.8
ENST00000422973.1
|
GALNT3
|
polypeptide N-acetylgalactosaminyltransferase 3
|
|
chr2_+_74002685
Show fit
|
0.21 |
ENST00000305799.8
|
TET3
|
tet methylcytosine dioxygenase 3
|
|
chr19_+_12995554
Show fit
|
0.20 |
ENST00000397661.6
|
NFIX
|
nuclear factor I X
|
|
chr9_-_20382461
Show fit
|
0.20 |
ENST00000380321.5
ENST00000629733.3
|
MLLT3
|
MLLT3 super elongation complex subunit
|
|
chr17_+_67825664
Show fit
|
0.20 |
ENST00000321892.8
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr2_-_26641363
Show fit
|
0.19 |
ENST00000288861.5
|
CIB4
|
calcium and integrin binding family member 4
|
|
chr17_-_82101850
Show fit
|
0.17 |
ENST00000583053.1
|
CCDC57
|
coiled-coil domain containing 57
|
|
chr17_+_67825494
Show fit
|
0.17 |
ENST00000306378.11
ENST00000544778.6
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr19_-_35490456
Show fit
|
0.16 |
ENST00000338897.4
ENST00000484218.6
|
KRTDAP
|
keratinocyte differentiation associated protein
|
|
chr11_+_63681444
Show fit
|
0.16 |
ENST00000341307.6
ENST00000356000.7
ENST00000542238.5
|
RTN3
|
reticulon 3
|
|
chr4_+_109560219
Show fit
|
0.16 |
ENST00000394650.7
|
MCUB
|
mitochondrial calcium uniporter dominant negative subunit beta
|
|
chr19_+_41350911
Show fit
|
0.16 |
ENST00000539627.5
|
TMEM91
|
transmembrane protein 91
|
|
chr6_+_155216637
Show fit
|
0.16 |
ENST00000275246.11
|
TIAM2
|
TIAM Rac1 associated GEF 2
|
|
chr11_+_63681483
Show fit
|
0.15 |
ENST00000339997.8
ENST00000540798.5
ENST00000545432.5
ENST00000543552.5
ENST00000377819.10
ENST00000537981.5
|
RTN3
|
reticulon 3
|
|
chr1_-_153615858
Show fit
|
0.15 |
ENST00000476873.5
|
S100A14
|
S100 calcium binding protein A14
|
|
chr19_+_35351552
Show fit
|
0.15 |
ENST00000246553.3
|
FFAR1
|
free fatty acid receptor 1
|
|
chr11_+_119149029
Show fit
|
0.13 |
ENST00000619701.5
|
ABCG4
|
ATP binding cassette subfamily G member 4
|
|
chr2_+_219572304
Show fit
|
0.13 |
ENST00000243786.3
|
INHA
|
inhibin subunit alpha
|
|
chr11_+_60429595
Show fit
|
0.13 |
ENST00000528905.1
ENST00000528093.1
|
MS4A5
|
membrane spanning 4-domains A5
|
|
chr11_+_111977300
Show fit
|
0.12 |
ENST00000615255.1
|
DIXDC1
|
DIX domain containing 1
|
|
chr14_+_103100328
Show fit
|
0.12 |
ENST00000559116.1
|
EXOC3L4
|
exocyst complex component 3 like 4
|
|
chr11_+_63681573
Show fit
|
0.12 |
ENST00000354497.4
|
RTN3
|
reticulon 3
|
|
chr4_-_20984011
Show fit
|
0.11 |
ENST00000382149.9
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4
|
|
chr8_-_143942524
Show fit
|
0.11 |
ENST00000357649.6
|
PLEC
|
plectin
|
|
chr12_-_91179472
Show fit
|
0.11 |
ENST00000550099.5
ENST00000546391.5
|
DCN
|
decorin
|
|
chr14_+_52314280
Show fit
|
0.11 |
ENST00000557436.1
ENST00000245457.6
|
PTGER2
|
prostaglandin E receptor 2
|
|
chr14_+_22086401
Show fit
|
0.11 |
ENST00000390451.2
|
TRAV23DV6
|
T cell receptor alpha variable 23/delta variable 6
|
|
chr16_-_14630200
Show fit
|
0.11 |
ENST00000650990.1
ENST00000651049.1
ENST00000652727.1
ENST00000652501.1
ENST00000539279.5
ENST00000651634.1
ENST00000651027.1
ENST00000420015.6
ENST00000651865.1
ENST00000437198.7
ENST00000341484.11
ENST00000650960.1
|
PARN
|
poly(A)-specific ribonuclease
|
|
chr3_-_185821092
Show fit
|
0.11 |
ENST00000421047.3
|
IGF2BP2
|
insulin like growth factor 2 mRNA binding protein 2
|
|
chr7_-_128405930
Show fit
|
0.10 |
ENST00000470772.5
ENST00000480861.5
ENST00000496200.5
|
IMPDH1
|
inosine monophosphate dehydrogenase 1
|
|
chr17_+_7888783
Show fit
|
0.10 |
ENST00000330494.12
ENST00000358181.8
|
CHD3
|
chromodomain helicase DNA binding protein 3
|
|
chr20_+_36876119
Show fit
|
0.10 |
ENST00000602922.5
ENST00000217320.8
|
TLDC2
|
TBC/LysM-associated domain containing 2
|
|
chr17_+_39200275
Show fit
|
0.10 |
ENST00000225430.9
|
RPL19
|
ribosomal protein L19
|
|
chr17_+_75754618
Show fit
|
0.10 |
ENST00000584939.1
|
ITGB4
|
integrin subunit beta 4
|
|
chr17_+_39200334
Show fit
|
0.10 |
ENST00000579260.5
ENST00000582193.5
|
RPL19
|
ribosomal protein L19
|
|
chr22_-_37580075
Show fit
|
0.09 |
ENST00000215886.6
|
LGALS2
|
galectin 2
|
|
chr19_+_35106510
Show fit
|
0.09 |
ENST00000648240.1
|
ENSG00000285526.1
|
novel protein
|
|
chr19_-_38735405
Show fit
|
0.09 |
ENST00000597987.5
ENST00000595177.1
|
CAPN12
|
calpain 12
|
|
chr17_+_39200302
Show fit
|
0.09 |
ENST00000579374.5
|
RPL19
|
ribosomal protein L19
|
|
chr4_-_156970903
Show fit
|
0.09 |
ENST00000422544.2
|
PDGFC
|
platelet derived growth factor C
|
|
chr19_-_35742431
Show fit
|
0.09 |
ENST00000592537.5
ENST00000246532.6
ENST00000588992.5
|
IGFLR1
|
IGF like family receptor 1
|
|
chr8_+_119873710
Show fit
|
0.09 |
ENST00000523492.5
ENST00000286234.6
|
DEPTOR
|
DEP domain containing MTOR interacting protein
|
|
chr17_-_76027212
Show fit
|
0.09 |
ENST00000586740.1
|
EVPL
|
envoplakin
|
|
chr1_-_51878799
Show fit
|
0.08 |
ENST00000354831.11
ENST00000544028.5
|
NRDC
|
nardilysin convertase
|
|
chr14_+_103100144
Show fit
|
0.08 |
ENST00000380069.7
|
EXOC3L4
|
exocyst complex component 3 like 4
|
|
chr1_+_27959943
Show fit
|
0.08 |
ENST00000675575.1
ENST00000373884.6
|
XKR8
|
XK related 8
|
|
chr17_+_74431338
Show fit
|
0.08 |
ENST00000342648.9
ENST00000652232.1
ENST00000481232.2
|
GPRC5C
|
G protein-coupled receptor class C group 5 member C
|
|
chr1_-_51878711
Show fit
|
0.08 |
ENST00000352171.12
|
NRDC
|
nardilysin convertase
|
|
chr19_+_41860236
Show fit
|
0.08 |
ENST00000601492.5
ENST00000600467.6
ENST00000598742.6
ENST00000221975.6
ENST00000598261.2
|
RPS19
|
ribosomal protein S19
|
|
chr12_-_31729010
Show fit
|
0.07 |
ENST00000537562.5
ENST00000537960.5
ENST00000281471.11
ENST00000536761.5
ENST00000542781.5
ENST00000457428.6
|
AMN1
|
antagonist of mitotic exit network 1 homolog
|
|
chr17_+_39200507
Show fit
|
0.07 |
ENST00000678573.1
|
RPL19
|
ribosomal protein L19
|
|
chr16_+_84145256
Show fit
|
0.07 |
ENST00000378553.10
|
DNAAF1
|
dynein axonemal assembly factor 1
|
|
chr1_+_16043736
Show fit
|
0.07 |
ENST00000619181.4
|
CLCNKB
|
chloride voltage-gated channel Kb
|
|
chr2_+_113406368
Show fit
|
0.07 |
ENST00000453673.3
|
IGKV1OR2-108
|
immunoglobulin kappa variable 1/OR2-108 (non-functional)
|
|
chr12_+_43758936
Show fit
|
0.06 |
ENST00000440781.6
ENST00000431837.5
ENST00000550616.5
ENST00000613694.5
ENST00000551736.5
|
IRAK4
|
interleukin 1 receptor associated kinase 4
|
|
chr13_+_50910018
Show fit
|
0.06 |
ENST00000645990.1
ENST00000642995.1
ENST00000636524.2
ENST00000643159.1
ENST00000645618.1
ENST00000611510.5
ENST00000642454.1
ENST00000646709.1
|
RNASEH2B
|
ribonuclease H2 subunit B
|
|
chr7_+_69967464
Show fit
|
0.06 |
ENST00000664521.1
|
AUTS2
|
activator of transcription and developmental regulator AUTS2
|
|
chr1_+_26159071
Show fit
|
0.06 |
ENST00000374268.5
|
FAM110D
|
family with sequence similarity 110 member D
|
|
chr14_-_89954659
Show fit
|
0.06 |
ENST00000555070.1
ENST00000316738.12
ENST00000538485.6
ENST00000556609.5
|
ENSG00000259053.1
EFCAB11
|
novel transcript
EF-hand calcium binding domain 11
|
|
chr11_+_66466745
Show fit
|
0.06 |
ENST00000349459.10
ENST00000320740.12
ENST00000524466.5
ENST00000526296.5
|
PELI3
|
pellino E3 ubiquitin protein ligase family member 3
|
|
chr2_+_69741974
Show fit
|
0.06 |
ENST00000409920.5
|
ANXA4
|
annexin A4
|
|
chr1_-_68232539
Show fit
|
0.06 |
ENST00000370976.7
ENST00000354777.6
|
WLS
|
Wnt ligand secretion mediator
|
|
chr19_+_43716095
Show fit
|
0.06 |
ENST00000596627.1
|
IRGC
|
immunity related GTPase cinema
|
|
chr3_+_136957948
Show fit
|
0.06 |
ENST00000329582.9
|
IL20RB
|
interleukin 20 receptor subunit beta
|
|
chr19_+_19516216
Show fit
|
0.05 |
ENST00000507754.9
ENST00000503283.5
ENST00000428459.6
ENST00000555938.1
|
NDUFA13
ENSG00000258674.5
|
NADH:ubiquinone oxidoreductase subunit A13
novel protein
|
|
chr2_+_219418369
Show fit
|
0.05 |
ENST00000373960.4
|
DES
|
desmin
|
|
chr10_-_89535575
Show fit
|
0.05 |
ENST00000371790.5
|
SLC16A12
|
solute carrier family 16 member 12
|
|
chr1_+_155173787
Show fit
|
0.05 |
ENST00000392451.6
ENST00000545012.5
ENST00000334634.9
ENST00000368385.8
ENST00000543729.5
ENST00000368383.7
ENST00000368382.5
|
TRIM46
|
tripartite motif containing 46
|
|
chr20_+_41136944
Show fit
|
0.05 |
ENST00000244007.7
|
PLCG1
|
phospholipase C gamma 1
|
|
chr17_-_76027296
Show fit
|
0.05 |
ENST00000301607.8
|
EVPL
|
envoplakin
|
|
chr1_-_68232514
Show fit
|
0.04 |
ENST00000262348.9
ENST00000370973.2
ENST00000370971.1
|
WLS
|
Wnt ligand secretion mediator
|
|
chr6_-_33289189
Show fit
|
0.04 |
ENST00000374617.9
|
WDR46
|
WD repeat domain 46
|
|
chr1_-_236065079
Show fit
|
0.04 |
ENST00000264187.7
ENST00000366595.7
|
NID1
|
nidogen 1
|
|
chr2_-_38602626
Show fit
|
0.04 |
ENST00000378915.7
|
HNRNPLL
|
heterogeneous nuclear ribonucleoprotein L like
|
|
chr17_+_56153458
Show fit
|
0.04 |
ENST00000318698.6
ENST00000682825.1
ENST00000566473.6
|
ANKFN1
|
ankyrin repeat and fibronectin type III domain containing 1
|
|
chrX_-_139642518
Show fit
|
0.04 |
ENST00000370573.8
ENST00000338585.6
|
MCF2
|
MCF.2 cell line derived transforming sequence
|
|
chr5_+_38258373
Show fit
|
0.04 |
ENST00000354891.7
|
EGFLAM
|
EGF like, fibronectin type III and laminin G domains
|
|
chr19_+_41114430
Show fit
|
0.04 |
ENST00000331105.7
|
CYP2F1
|
cytochrome P450 family 2 subfamily F member 1
|
|
chr5_+_38258551
Show fit
|
0.04 |
ENST00000322350.10
|
EGFLAM
|
EGF like, fibronectin type III and laminin G domains
|
|
chr17_+_32928126
Show fit
|
0.03 |
ENST00000579849.6
ENST00000578289.5
ENST00000439138.5
|
TMEM98
|
transmembrane protein 98
|
|
chr17_-_44123628
Show fit
|
0.03 |
ENST00000587135.1
ENST00000225983.10
ENST00000682912.1
ENST00000588703.5
|
HDAC5
|
histone deacetylase 5
|
|
chr1_-_34859717
Show fit
|
0.03 |
ENST00000423898.1
ENST00000521580.3
ENST00000456842.1
|
SMIM12
|
small integral membrane protein 12
|
|
chr1_+_156815633
Show fit
|
0.03 |
ENST00000392302.7
ENST00000674537.1
|
NTRK1
|
neurotrophic receptor tyrosine kinase 1
|
|
chr17_+_45241067
Show fit
|
0.03 |
ENST00000587489.5
|
FMNL1
|
formin like 1
|
|
chr17_-_40867200
Show fit
|
0.03 |
ENST00000647902.1
ENST00000251643.5
|
KRT12
|
keratin 12
|
|
chr3_-_48088824
Show fit
|
0.03 |
ENST00000439356.2
ENST00000395734.7
ENST00000426837.6
|
MAP4
|
microtubule associated protein 4
|
|
chr10_-_31928864
Show fit
|
0.03 |
ENST00000375245.8
ENST00000396144.8
|
ARHGAP12
|
Rho GTPase activating protein 12
|
|
chr18_-_55586092
Show fit
|
0.03 |
ENST00000563888.6
ENST00000540999.5
ENST00000627685.2
|
TCF4
|
transcription factor 4
|
|
chr12_+_113057687
Show fit
|
0.02 |
ENST00000257600.3
|
DTX1
|
deltex E3 ubiquitin ligase 1
|
|
chr11_+_60429567
Show fit
|
0.02 |
ENST00000300190.7
|
MS4A5
|
membrane spanning 4-domains A5
|
|
chr10_-_95441015
Show fit
|
0.02 |
ENST00000371241.5
ENST00000354106.7
ENST00000371239.5
ENST00000361941.7
ENST00000277982.9
ENST00000371245.7
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr16_-_636270
Show fit
|
0.02 |
ENST00000397665.6
ENST00000397666.6
ENST00000301686.13
ENST00000338401.8
ENST00000397664.8
ENST00000568830.1
ENST00000614890.4
|
METTL26
|
methyltransferase like 26
|
|
chr15_+_74174403
Show fit
|
0.02 |
ENST00000560862.1
ENST00000395118.1
|
ISLR
|
immunoglobulin superfamily containing leucine rich repeat
|
|
chr22_+_39570753
Show fit
|
0.02 |
ENST00000407673.5
ENST00000401624.5
ENST00000404898.5
ENST00000402142.4
|
CACNA1I
|
calcium voltage-gated channel subunit alpha1 I
|
|
chr16_+_74999003
Show fit
|
0.02 |
ENST00000335325.9
ENST00000320619.10
|
ZNRF1
|
zinc and ring finger 1
|
|
chr20_-_59940289
Show fit
|
0.02 |
ENST00000370996.5
|
PPP1R3D
|
protein phosphatase 1 regulatory subunit 3D
|
|
chrX_+_73447042
Show fit
|
0.01 |
ENST00000373514.3
|
CDX4
|
caudal type homeobox 4
|
|
chr4_+_105710809
Show fit
|
0.01 |
ENST00000360505.9
ENST00000510865.5
ENST00000509336.5
|
GSTCD
|
glutathione S-transferase C-terminal domain containing
|
|
chr10_-_31928790
Show fit
|
0.01 |
ENST00000375250.9
ENST00000344936.7
|
ARHGAP12
|
Rho GTPase activating protein 12
|
|
chr18_-_4455257
Show fit
|
0.01 |
ENST00000581527.5
|
DLGAP1
|
DLG associated protein 1
|
|
chr16_-_8797624
Show fit
|
0.01 |
ENST00000333050.7
|
TMEM186
|
transmembrane protein 186
|
|
chr9_+_35673917
Show fit
|
0.01 |
ENST00000617161.1
ENST00000378357.9
|
CA9
|
carbonic anhydrase 9
|
|
chr3_-_69386079
Show fit
|
0.01 |
ENST00000398540.8
|
FRMD4B
|
FERM domain containing 4B
|
|
chr8_+_144504131
Show fit
|
0.01 |
ENST00000394955.3
|
GPT
|
glutamic--pyruvic transaminase
|
|
chr5_+_139561159
Show fit
|
0.01 |
ENST00000505007.5
|
UBE2D2
|
ubiquitin conjugating enzyme E2 D2
|
|
chr3_+_48440246
Show fit
|
0.01 |
ENST00000438607.2
|
TMA7
|
translation machinery associated 7 homolog
|
|
chr12_-_56488153
Show fit
|
0.00 |
ENST00000311966.9
|
GLS2
|
glutaminase 2
|
|
chr20_-_54173516
Show fit
|
0.00 |
ENST00000395955.7
|
CYP24A1
|
cytochrome P450 family 24 subfamily A member 1
|
|
chrX_-_139642889
Show fit
|
0.00 |
ENST00000370576.9
|
MCF2
|
MCF.2 cell line derived transforming sequence
|
|
chr11_+_237016
Show fit
|
0.00 |
ENST00000352303.9
|
PSMD13
|
proteasome 26S subunit, non-ATPase 13
|
|
chr2_+_89884740
Show fit
|
0.00 |
ENST00000509129.1
|
IGKV1D-37
|
immunoglobulin kappa variable 1D-37 (non-functional)
|
|
chr6_+_31547560
Show fit
|
0.00 |
ENST00000376148.9
ENST00000376145.8
|
NFKBIL1
|
NFKB inhibitor like 1
|
|
chr1_-_182672882
Show fit
|
0.00 |
ENST00000367557.8
ENST00000258302.8
|
RGS8
|
regulator of G protein signaling 8
|
|
chr16_+_89918855
Show fit
|
0.00 |
ENST00000555147.2
|
MC1R
|
melanocortin 1 receptor
|
|
chrX_-_139642835
Show fit
|
0.00 |
ENST00000536274.5
|
MCF2
|
MCF.2 cell line derived transforming sequence
|
|
chr14_+_21749163
Show fit
|
0.00 |
ENST00000390427.3
|
TRAV5
|
T cell receptor alpha variable 5
|