Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for UUGGCAC
Z-value: 0.36
miRNA associated with seed UUGGCAC
| Name | miRBASE accession |
|---|---|
|
hsa-miR-1271-5p
|
MIMAT0005796 |
|
hsa-miR-96-5p
|
MIMAT0000095 |
Activity profile of UUGGCAC motif
Sorted Z-values of UUGGCAC motif
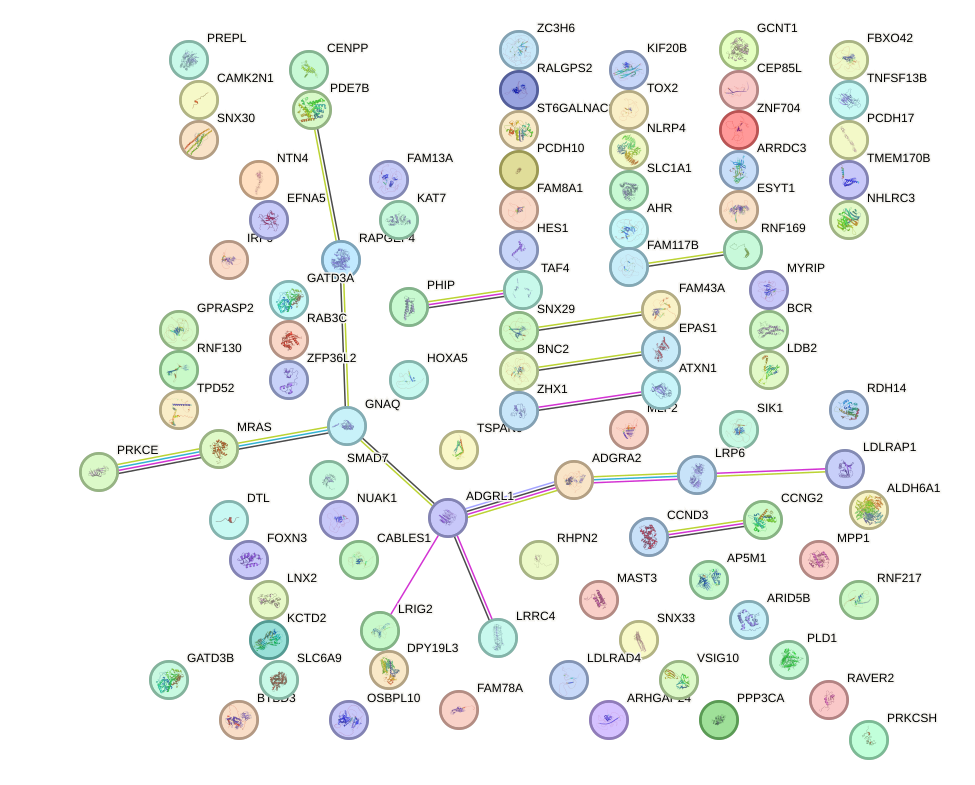
Network of associatons between targets according to the STRING database.
First level regulatory network of UUGGCAC
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_76459152 | 0.84 |
ENST00000444201.6
ENST00000376730.5 |
GCNT1
|
glucosaminyl (N-acetyl) transferase 1 |
| chr4_-_16898619 | 0.77 |
ENST00000502640.5
ENST00000304523.10 ENST00000506732.1 |
LDB2
|
LIM domain binding 2 |
| chr13_+_57631735 | 0.72 |
ENST00000377918.8
|
PCDH17
|
protocadherin 17 |
| chr18_+_23135452 | 0.69 |
ENST00000580153.5
ENST00000256925.12 |
CABLES1
|
Cdk5 and Abl enzyme substrate 1 |
| chr3_+_194136138 | 0.69 |
ENST00000232424.4
|
HES1
|
hes family bHLH transcription factor 1 |
| chr9_-_131270493 | 0.58 |
ENST00000372269.7
ENST00000464831.1 |
FAM78A
|
family with sequence similarity 78 member A |
| chr3_+_39809602 | 0.57 |
ENST00000302541.11
ENST00000396217.7 |
MYRIP
|
myosin VIIA and Rab interacting protein |
| chr8_+_37796906 | 0.51 |
ENST00000315215.11
|
ADGRA2
|
adhesion G protein-coupled receptor A2 |
| chr12_-_106138946 | 0.51 |
ENST00000261402.7
|
NUAK1
|
NUAK family kinase 1 |
| chr7_+_17298642 | 0.46 |
ENST00000242057.9
|
AHR
|
aryl hydrocarbon receptor |
| chr18_-_48950960 | 0.45 |
ENST00000262158.8
|
SMAD7
|
SMAD family member 7 |
| chr17_+_49788672 | 0.45 |
ENST00000454930.6
ENST00000259021.9 ENST00000509773.5 ENST00000510819.5 ENST00000424009.6 |
KAT7
|
lysine acetyltransferase 7 |
| chr6_-_16761447 | 0.44 |
ENST00000244769.8
ENST00000436367.6 |
ATXN1
|
ataxin 1 |
| chr20_+_43916142 | 0.44 |
ENST00000423191.6
ENST00000372999.5 |
TOX2
|
TOX high mobility group box family member 2 |
| chrX_+_102712438 | 0.44 |
ENST00000486814.2
ENST00000535209.6 ENST00000543253.6 ENST00000332262.10 ENST00000483720.6 |
GPRASP2
|
G protein-coupled receptor associated sorting protein 2 |
| chr9_+_112750722 | 0.40 |
ENST00000374232.8
|
SNX30
|
sorting nexin family member 30 |
| chr2_+_45651650 | 0.38 |
ENST00000306156.8
|
PRKCE
|
protein kinase C epsilon |
| chr4_+_133149278 | 0.38 |
ENST00000264360.7
|
PCDH10
|
protocadherin 10 |
| chr6_+_11537738 | 0.36 |
ENST00000379426.2
|
TMEM170B
|
transmembrane protein 170B |
| chr4_-_101347471 | 0.36 |
ENST00000323055.10
ENST00000512215.5 |
PPP3CA
|
protein phosphatase 3 catalytic subunit alpha |
| chr1_+_64745089 | 0.36 |
ENST00000294428.7
ENST00000371072.8 |
RAVER2
|
ribonucleoprotein, PTB binding 2 |
| chr13_-_27620520 | 0.35 |
ENST00000316334.5
|
LNX2
|
ligand of numb-protein X 2 |
| chr7_-_27143672 | 0.35 |
ENST00000222726.4
|
HOXA5
|
homeobox A5 |
| chr2_+_172735912 | 0.35 |
ENST00000409036.5
|
RAPGEF4
|
Rap guanine nucleotide exchange factor 4 |
| chr12_-_95790755 | 0.34 |
ENST00000343702.9
ENST00000344911.8 |
NTN4
|
netrin 4 |
| chr8_-_80171496 | 0.33 |
ENST00000379096.9
ENST00000518937.6 |
TPD52
|
tumor protein D52 |
| chr1_+_76074698 | 0.32 |
ENST00000328299.4
|
ST6GALNAC3
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr6_-_41941795 | 0.32 |
ENST00000372991.9
|
CCND3
|
cyclin D3 |
| chr6_+_135851681 | 0.32 |
ENST00000308191.11
|
PDE7B
|
phosphodiesterase 7B |
| chr9_-_16870662 | 0.31 |
ENST00000380672.9
|
BNC2
|
basonuclin 2 |
| chr14_+_57268963 | 0.30 |
ENST00000261558.8
|
AP5M1
|
adaptor related protein complex 5 subunit mu 1 |
| chr12_-_118103998 | 0.30 |
ENST00000359236.10
|
VSIG10
|
V-set and immunoglobulin domain containing 10 |
| chr9_+_4490388 | 0.29 |
ENST00000262352.8
|
SLC1A1
|
solute carrier family 1 member 1 |
| chr12_+_32502114 | 0.29 |
ENST00000682739.1
ENST00000427716.7 ENST00000583694.2 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr1_+_25543598 | 0.29 |
ENST00000374338.5
|
LDLRAP1
|
low density lipoprotein receptor adaptor protein 1 |
| chr19_+_32405789 | 0.28 |
ENST00000586987.5
|
DPY19L3
|
dpy-19 like C-mannosyltransferase 3 |
| chr2_-_43226594 | 0.28 |
ENST00000282388.4
|
ZFP36L2
|
ZFP36 ring finger protein like 2 |
| chr1_-_16352420 | 0.28 |
ENST00000375592.8
|
FBXO42
|
F-box protein 42 |
| chr2_+_202634960 | 0.27 |
ENST00000392238.3
|
FAM117B
|
family with sequence similarity 117 member B |
| chr12_-_56333693 | 0.26 |
ENST00000425394.7
ENST00000548043.5 |
PAN2
|
poly(A) specific ribonuclease subunit PAN2 |
| chr20_+_11890723 | 0.26 |
ENST00000254977.7
|
BTBD3
|
BTB domain containing 3 |
| chr1_-_209806124 | 0.26 |
ENST00000367021.8
ENST00000542854.5 |
IRF6
|
interferon regulatory factor 6 |
| chr8_-_80874771 | 0.26 |
ENST00000327835.7
|
ZNF704
|
zinc finger protein 704 |
| chr3_+_194685874 | 0.25 |
ENST00000329759.6
|
FAM43A
|
family with sequence similarity 43 member A |
| chr9_+_128068172 | 0.25 |
ENST00000373068.6
ENST00000373069.10 |
SLC25A25
|
solute carrier family 25 member 25 |
| chr20_-_62065834 | 0.23 |
ENST00000252996.9
|
TAF4
|
TATA-box binding protein associated factor 4 |
| chr1_-_44031446 | 0.23 |
ENST00000372310.8
ENST00000466926.1 |
SLC6A9
|
solute carrier family 6 member 9 |
| chr5_-_107670897 | 0.23 |
ENST00000333274.11
|
EFNA5
|
ephrin A5 |
| chr22_+_23180365 | 0.23 |
ENST00000359540.7
ENST00000305877.13 |
BCR
|
BCR activator of RhoGEF and GTPase |
| chr12_-_12267003 | 0.22 |
ENST00000535731.1
ENST00000261349.9 |
LRP6
|
LDL receptor related protein 6 |
| chr21_-_43427131 | 0.21 |
ENST00000270162.8
|
SIK1
|
salt inducible kinase 1 |
| chr19_-_14206168 | 0.21 |
ENST00000361434.7
ENST00000340736.10 |
ADGRL1
|
adhesion G protein-coupled receptor L1 |
| chr12_+_3077355 | 0.21 |
ENST00000537971.5
ENST00000011898.10 ENST00000649909.1 |
TSPAN9
|
tetraspanin 9 |
| chr1_+_178725227 | 0.21 |
ENST00000367635.8
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr12_+_32107151 | 0.21 |
ENST00000548411.5
|
BICD1
|
BICD cargo adaptor 1 |
| chr19_-_33064872 | 0.21 |
ENST00000254260.8
|
RHPN2
|
rhophilin Rho GTPase binding protein 2 |
| chr6_+_124962420 | 0.21 |
ENST00000521654.7
ENST00000560949.5 |
RNF217
|
ring finger protein 217 |
| chr18_+_13218769 | 0.21 |
ENST00000677055.1
ENST00000399848.7 |
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr9_-_78031775 | 0.21 |
ENST00000286548.9
|
GNAQ
|
G protein subunit alpha q |
| chr10_+_61901678 | 0.21 |
ENST00000644638.1
ENST00000681100.1 ENST00000279873.12 |
ARID5B
|
AT-rich interaction domain 5B |
| chr13_+_39038292 | 0.20 |
ENST00000470258.5
|
NHLRC3
|
NHL repeat containing 3 |
| chr16_+_11976709 | 0.20 |
ENST00000566228.6
|
SNX29
|
sorting nexin 29 |
| chr6_+_17600273 | 0.20 |
ENST00000259963.4
|
FAM8A1
|
family with sequence similarity 8 member A1 |
| chr7_-_128031422 | 0.20 |
ENST00000249363.4
|
LRRC4
|
leucine rich repeat containing 4 |
| chr17_+_75047205 | 0.20 |
ENST00000322444.7
|
KCTD2
|
potassium channel tetramerization domain containing 2 |
| chr3_-_31981228 | 0.20 |
ENST00000396556.7
ENST00000438237.6 |
OSBPL10
|
oxysterol binding protein like 10 |
| chr3_-_171809770 | 0.20 |
ENST00000331659.2
|
PLD1
|
phospholipase D1 |
| chr2_-_44361754 | 0.19 |
ENST00000409272.5
ENST00000410081.5 ENST00000541738.5 |
PREPL
|
prolyl endopeptidase like |
| chr6_-_79078247 | 0.19 |
ENST00000275034.5
|
PHIP
|
pleckstrin homology domain interacting protein |
| chr14_-_74084393 | 0.19 |
ENST00000350259.8
ENST00000553458.6 |
ALDH6A1
|
aldehyde dehydrogenase 6 family member A1 |
| chr1_+_212035717 | 0.19 |
ENST00000366991.5
|
DTL
|
denticleless E3 ubiquitin protein ligase homolog |
| chr1_-_20486197 | 0.18 |
ENST00000375078.4
|
CAMK2N1
|
calcium/calmodulin dependent protein kinase II inhibitor 1 |
| chr4_+_77157189 | 0.18 |
ENST00000316355.10
ENST00000502280.5 |
CCNG2
|
cyclin G2 |
| chr2_+_46297397 | 0.18 |
ENST00000263734.5
|
EPAS1
|
endothelial PAS domain protein 1 |
| chr3_+_138347648 | 0.18 |
ENST00000614350.4
ENST00000289104.8 |
MRAS
|
muscle RAS oncogene homolog |
| chr9_+_92325910 | 0.18 |
ENST00000375587.8
ENST00000618653.1 |
CENPP
|
centromere protein P |
| chr4_+_85475131 | 0.18 |
ENST00000395184.6
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chrX_-_154805386 | 0.17 |
ENST00000393531.5
ENST00000369534.8 ENST00000453245.5 ENST00000428488.1 ENST00000369531.1 |
MPP1
|
membrane palmitoylated protein 1 |
| chr11_+_74748831 | 0.17 |
ENST00000299563.5
|
RNF169
|
ring finger protein 169 |
| chr1_+_81800368 | 0.17 |
ENST00000674489.1
ENST00000674442.1 ENST00000674419.1 ENST00000674407.1 ENST00000674168.1 ENST00000674307.1 ENST00000674209.1 ENST00000370715.5 ENST00000370713.5 ENST00000319517.10 ENST00000627151.2 ENST00000370717.6 |
ADGRL2
|
adhesion G protein-coupled receptor L2 |
| chr14_-_89619118 | 0.17 |
ENST00000345097.8
ENST00000555855.5 ENST00000555353.5 |
FOXN3
|
forkhead box N3 |
| chr8_-_123274255 | 0.17 |
ENST00000622816.2
ENST00000395571.8 |
ZHX1-C8orf76
ZHX1
|
ZHX1-C8orf76 readthrough zinc fingers and homeoboxes 1 |
| chr5_+_58583068 | 0.17 |
ENST00000282878.6
|
RAB3C
|
RAB3C, member RAS oncogene family |
| chr5_-_91383310 | 0.17 |
ENST00000265138.4
|
ARRDC3
|
arrestin domain containing 3 |
| chr4_-_88823306 | 0.16 |
ENST00000395002.6
|
FAM13A
|
family with sequence similarity 13 member A |
| chr19_+_18097763 | 0.16 |
ENST00000262811.10
|
MAST3
|
microtubule associated serine/threonine kinase 3 |
| chr12_+_56128217 | 0.16 |
ENST00000267113.4
ENST00000394048.10 |
ESYT1
|
extended synaptotagmin 1 |
| chr6_-_118651522 | 0.16 |
ENST00000368491.8
|
CEP85L
|
centrosomal protein 85 like |
| chr1_+_113073162 | 0.16 |
ENST00000361127.6
|
LRIG2
|
leucine rich repeats and immunoglobulin like domains 2 |
| chr7_-_87059639 | 0.16 |
ENST00000450689.7
|
ELAPOR2
|
endosome-lysosome associated apoptosis and autophagy regulator family member 2 |
| chr2_+_112275588 | 0.16 |
ENST00000409871.6
ENST00000343936.4 |
ZC3H6
|
zinc finger CCCH-type containing 6 |
| chr5_-_180072086 | 0.15 |
ENST00000261947.4
|
RNF130
|
ring finger protein 130 |
| chr1_+_201829132 | 0.15 |
ENST00000361565.9
|
IPO9
|
importin 9 |
| chr8_+_28494190 | 0.15 |
ENST00000537916.2
ENST00000240093.8 ENST00000523546.1 |
FZD3
|
frizzled class receptor 3 |
| chr13_+_99981775 | 0.15 |
ENST00000376335.8
|
ZIC2
|
Zic family member 2 |
| chr1_-_114670018 | 0.15 |
ENST00000393274.6
ENST00000393276.7 |
DENND2C
|
DENN domain containing 2C |
| chr4_-_176002332 | 0.15 |
ENST00000280187.11
ENST00000512509.5 |
GPM6A
|
glycoprotein M6A |
| chr6_-_32190170 | 0.15 |
ENST00000375050.6
|
PBX2
|
PBX homeobox 2 |
| chr3_-_134374439 | 0.15 |
ENST00000513145.1
ENST00000249883.10 ENST00000422605.6 |
AMOTL2
|
angiomotin like 2 |
| chr1_-_154961720 | 0.15 |
ENST00000368457.3
|
PYGO2
|
pygopus family PHD finger 2 |
| chr11_-_95231046 | 0.14 |
ENST00000416495.6
ENST00000536441.7 |
SESN3
|
sestrin 3 |
| chr1_-_40665654 | 0.14 |
ENST00000372684.8
|
RIMS3
|
regulating synaptic membrane exocytosis 3 |
| chr17_-_59892708 | 0.14 |
ENST00000346141.10
|
TUBD1
|
tubulin delta 1 |
| chr12_-_57520480 | 0.14 |
ENST00000642841.1
ENST00000547303.5 ENST00000552740.5 ENST00000547526.1 ENST00000346473.8 ENST00000551116.5 |
ENSG00000285133.1
DDIT3
|
novel protein DNA damage inducible transcript 3 |
| chr19_-_6279921 | 0.14 |
ENST00000252674.9
|
MLLT1
|
MLLT1 super elongation complex subunit |
| chr12_+_12891554 | 0.14 |
ENST00000014914.6
|
GPRC5A
|
G protein-coupled receptor class C group 5 member A |
| chr2_+_108588453 | 0.14 |
ENST00000393310.5
|
LIMS1
|
LIM zinc finger domain containing 1 |
| chr18_-_75209126 | 0.14 |
ENST00000322342.4
|
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2 |
| chr11_-_64803152 | 0.14 |
ENST00000439069.5
ENST00000294066.7 ENST00000377350.7 |
MAP4K2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr16_+_77788554 | 0.14 |
ENST00000302536.3
|
VAT1L
|
vesicle amine transport 1 like |
| chr9_-_113299196 | 0.14 |
ENST00000441031.3
|
RNF183
|
ring finger protein 183 |
| chr2_-_157874976 | 0.14 |
ENST00000682025.1
ENST00000683487.1 ENST00000682300.1 ENST00000683441.1 ENST00000684595.1 ENST00000683426.1 ENST00000683820.1 ENST00000263640.7 |
ACVR1
|
activin A receptor type 1 |
| chr13_-_40666600 | 0.14 |
ENST00000379561.6
|
FOXO1
|
forkhead box O1 |
| chr6_+_35213948 | 0.14 |
ENST00000274938.8
|
SCUBE3
|
signal peptide, CUB domain and EGF like domain containing 3 |
| chr6_+_52362088 | 0.14 |
ENST00000635984.1
ENST00000635760.1 ENST00000442253.3 |
EFHC1
PAQR8
|
EF-hand domain containing 1 progestin and adipoQ receptor family member 8 |
| chr17_+_63622406 | 0.13 |
ENST00000579585.5
ENST00000361733.8 ENST00000584573.5 ENST00000361357.7 |
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr3_-_39153512 | 0.13 |
ENST00000273153.10
|
CSRNP1
|
cysteine and serine rich nuclear protein 1 |
| chr8_-_81842192 | 0.13 |
ENST00000353788.8
ENST00000520618.5 ENST00000518183.5 ENST00000396330.6 ENST00000519119.5 |
SNX16
|
sorting nexin 16 |
| chr11_+_134224610 | 0.13 |
ENST00000281187.10
ENST00000525095.2 |
VPS26B
|
VPS26, retromer complex component B |
| chr3_-_171460368 | 0.13 |
ENST00000436636.7
ENST00000465393.1 ENST00000341852.10 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr17_+_74326190 | 0.13 |
ENST00000551294.5
ENST00000389916.5 |
KIF19
|
kinesin family member 19 |
| chr14_-_39432414 | 0.13 |
ENST00000554932.1
ENST00000298097.7 |
FBXO33
|
F-box protein 33 |
| chrX_-_19887585 | 0.13 |
ENST00000397821.8
|
SH3KBP1
|
SH3 domain containing kinase binding protein 1 |
| chr3_+_179653032 | 0.13 |
ENST00000680587.1
ENST00000681064.1 ENST00000263966.8 ENST00000681358.1 ENST00000679749.1 |
USP13
|
ubiquitin specific peptidase 13 |
| chr15_-_72197772 | 0.13 |
ENST00000309731.12
|
GRAMD2A
|
GRAM domain containing 2A |
| chr2_-_99489955 | 0.13 |
ENST00000393445.7
ENST00000258428.8 |
REV1
|
REV1 DNA directed polymerase |
| chr10_+_110644306 | 0.13 |
ENST00000369519.4
|
RBM20
|
RNA binding motif protein 20 |
| chr12_-_57237090 | 0.13 |
ENST00000556732.1
|
NDUFA4L2
|
NDUFA4 mitochondrial complex associated like 2 |
| chr1_-_109041986 | 0.13 |
ENST00000400794.7
ENST00000528747.1 ENST00000361054.7 |
WDR47
|
WD repeat domain 47 |
| chr1_+_3069160 | 0.13 |
ENST00000511072.5
|
PRDM16
|
PR/SET domain 16 |
| chr3_-_197555950 | 0.13 |
ENST00000445160.2
ENST00000392379.6 ENST00000446746.5 ENST00000432819.5 ENST00000441275.5 ENST00000392378.6 |
BDH1
|
3-hydroxybutyrate dehydrogenase 1 |
| chr4_-_42657085 | 0.12 |
ENST00000264449.14
ENST00000510289.1 ENST00000381668.9 |
ATP8A1
|
ATPase phospholipid transporting 8A1 |
| chr1_-_54887161 | 0.12 |
ENST00000535035.6
ENST00000371269.9 ENST00000436604.2 |
DHCR24
|
24-dehydrocholesterol reductase |
| chr7_+_87345656 | 0.12 |
ENST00000331536.8
ENST00000419147.6 ENST00000412227.6 |
CROT
|
carnitine O-octanoyltransferase |
| chr6_+_42782020 | 0.12 |
ENST00000314073.9
|
BICRAL
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr13_+_114314474 | 0.12 |
ENST00000463003.2
ENST00000645174.1 ENST00000361283.4 ENST00000644294.1 |
CHAMP1
|
chromosome alignment maintaining phosphoprotein 1 |
| chr1_+_28369705 | 0.12 |
ENST00000373839.8
|
PHACTR4
|
phosphatase and actin regulator 4 |
| chr3_-_56468346 | 0.12 |
ENST00000288221.11
|
ERC2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr8_+_73294594 | 0.12 |
ENST00000240285.10
|
RDH10
|
retinol dehydrogenase 10 |
| chr12_-_42144823 | 0.12 |
ENST00000398675.8
|
GXYLT1
|
glucoside xylosyltransferase 1 |
| chr2_-_217944005 | 0.12 |
ENST00000611415.4
ENST00000615025.5 ENST00000449814.1 ENST00000171887.8 |
TNS1
|
tensin 1 |
| chr6_-_6320642 | 0.12 |
ENST00000451619.1
ENST00000264870.8 |
F13A1
|
coagulation factor XIII A chain |
| chr14_+_63204436 | 0.12 |
ENST00000316754.8
|
RHOJ
|
ras homolog family member J |
| chr1_+_22451843 | 0.12 |
ENST00000375647.5
ENST00000404138.5 ENST00000374651.8 ENST00000400239.6 |
ZBTB40
|
zinc finger and BTB domain containing 40 |
| chr2_+_218399838 | 0.12 |
ENST00000273062.7
|
CTDSP1
|
CTD small phosphatase 1 |
| chr4_-_16226460 | 0.12 |
ENST00000405303.7
|
TAPT1
|
transmembrane anterior posterior transformation 1 |
| chr8_+_38974212 | 0.12 |
ENST00000302495.5
|
HTRA4
|
HtrA serine peptidase 4 |
| chr7_-_134316912 | 0.12 |
ENST00000378509.9
|
SLC35B4
|
solute carrier family 35 member B4 |
| chr12_+_59689337 | 0.11 |
ENST00000261187.8
|
SLC16A7
|
solute carrier family 16 member 7 |
| chr1_-_52552994 | 0.11 |
ENST00000355809.4
ENST00000528642.5 ENST00000470626.1 ENST00000257177.9 ENST00000371544.7 |
TUT4
|
terminal uridylyl transferase 4 |
| chr2_-_150487658 | 0.11 |
ENST00000375734.6
ENST00000263895.9 ENST00000454202.5 |
RND3
|
Rho family GTPase 3 |
| chr13_-_20161038 | 0.11 |
ENST00000241125.4
|
GJA3
|
gap junction protein alpha 3 |
| chr22_-_19178719 | 0.11 |
ENST00000215882.10
|
SLC25A1
|
solute carrier family 25 member 1 |
| chr8_+_96493803 | 0.11 |
ENST00000518385.5
ENST00000302190.9 |
SDC2
|
syndecan 2 |
| chr5_-_39074377 | 0.11 |
ENST00000514735.1
ENST00000357387.8 ENST00000296782.9 |
RICTOR
|
RPTOR independent companion of MTOR complex 2 |
| chr3_-_86991135 | 0.11 |
ENST00000398399.7
|
VGLL3
|
vestigial like family member 3 |
| chr10_+_35247015 | 0.11 |
ENST00000490012.6
ENST00000374706.5 ENST00000493157.6 |
CCNY
|
cyclin Y |
| chr7_+_107168961 | 0.11 |
ENST00000468410.5
ENST00000478930.5 ENST00000464009.1 ENST00000222574.9 |
HBP1
|
HMG-box transcription factor 1 |
| chr11_-_73598183 | 0.11 |
ENST00000064778.8
|
FAM168A
|
family with sequence similarity 168 member A |
| chr16_+_24729641 | 0.11 |
ENST00000395799.8
|
TNRC6A
|
trinucleotide repeat containing adaptor 6A |
| chr10_-_86366784 | 0.10 |
ENST00000327946.12
|
GRID1
|
glutamate ionotropic receptor delta type subunit 1 |
| chr4_-_77819356 | 0.10 |
ENST00000649644.1
ENST00000504123.6 ENST00000515441.2 |
CNOT6L
|
CCR4-NOT transcription complex subunit 6 like |
| chr16_-_23510389 | 0.10 |
ENST00000562117.1
ENST00000567468.5 ENST00000562944.5 ENST00000309859.8 |
GGA2
|
golgi associated, gamma adaptin ear containing, ARF binding protein 2 |
| chr5_+_87268922 | 0.10 |
ENST00000456692.6
ENST00000512763.5 ENST00000506290.1 |
RASA1
|
RAS p21 protein activator 1 |
| chr2_-_100417608 | 0.10 |
ENST00000264249.8
|
CHST10
|
carbohydrate sulfotransferase 10 |
| chr4_+_144646145 | 0.10 |
ENST00000296575.8
ENST00000434550.2 |
HHIP
|
hedgehog interacting protein |
| chr2_+_161416273 | 0.10 |
ENST00000389554.8
|
TBR1
|
T-box brain transcription factor 1 |
| chr7_+_107470050 | 0.10 |
ENST00000304402.6
|
GPR22
|
G protein-coupled receptor 22 |
| chr6_+_87472925 | 0.10 |
ENST00000369556.7
ENST00000369557.9 ENST00000369552.9 |
SLC35A1
|
solute carrier family 35 member A1 |
| chr22_+_30635746 | 0.10 |
ENST00000343605.5
|
SLC35E4
|
solute carrier family 35 member E4 |
| chr22_+_20965108 | 0.10 |
ENST00000399167.6
ENST00000399163.6 |
AIFM3
|
apoptosis inducing factor mitochondria associated 3 |
| chr1_-_153963505 | 0.10 |
ENST00000356205.9
ENST00000537590.5 |
SLC39A1
|
solute carrier family 39 member 1 |
| chr8_-_94949350 | 0.10 |
ENST00000448464.6
ENST00000342697.5 |
TP53INP1
|
tumor protein p53 inducible nuclear protein 1 |
| chr4_+_92303946 | 0.10 |
ENST00000282020.9
|
GRID2
|
glutamate ionotropic receptor delta type subunit 2 |
| chr6_-_98947911 | 0.10 |
ENST00000369244.7
ENST00000229971.2 |
FBXL4
|
F-box and leucine rich repeat protein 4 |
| chr2_+_134120169 | 0.09 |
ENST00000409645.5
|
MGAT5
|
alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase |
| chr16_-_47143934 | 0.09 |
ENST00000562435.6
|
NETO2
|
neuropilin and tolloid like 2 |
| chr2_+_96816236 | 0.09 |
ENST00000377060.7
ENST00000305510.4 |
CNNM3
|
cyclin and CBS domain divalent metal cation transport mediator 3 |
| chr19_+_10625507 | 0.09 |
ENST00000590857.5
ENST00000588688.5 ENST00000586078.5 ENST00000335757.10 |
SLC44A2
|
solute carrier family 44 member 2 |
| chr9_+_2621766 | 0.09 |
ENST00000382100.8
|
VLDLR
|
very low density lipoprotein receptor |
| chr22_-_46537593 | 0.09 |
ENST00000262738.9
ENST00000674500.2 |
CELSR1
|
cadherin EGF LAG seven-pass G-type receptor 1 |
| chr8_-_123541197 | 0.09 |
ENST00000517956.5
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr10_-_60944132 | 0.09 |
ENST00000337910.10
|
RHOBTB1
|
Rho related BTB domain containing 1 |
| chr2_+_240435652 | 0.09 |
ENST00000264039.7
|
GPC1
|
glypican 1 |
| chrX_-_107716401 | 0.09 |
ENST00000486554.1
ENST00000372390.8 |
TSC22D3
|
TSC22 domain family member 3 |
| chr12_-_108731505 | 0.09 |
ENST00000261401.8
ENST00000552871.5 |
CORO1C
|
coronin 1C |
| chr2_-_183038405 | 0.09 |
ENST00000361354.9
|
NCKAP1
|
NCK associated protein 1 |
| chr3_-_56801939 | 0.09 |
ENST00000296315.8
ENST00000495373.5 |
ARHGEF3
|
Rho guanine nucleotide exchange factor 3 |
| chr17_-_43022350 | 0.09 |
ENST00000587173.5
ENST00000355653.8 |
VAT1
|
vesicle amine transport 1 |
| chr3_-_115071333 | 0.09 |
ENST00000462705.5
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr12_+_14365661 | 0.09 |
ENST00000261168.9
ENST00000538511.5 ENST00000545723.1 ENST00000543189.5 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr22_-_36507022 | 0.09 |
ENST00000216187.10
ENST00000397224.9 ENST00000423980.1 |
FOXRED2
|
FAD dependent oxidoreductase domain containing 2 |
| chr12_+_57550027 | 0.09 |
ENST00000674619.1
ENST00000676359.1 ENST00000286452.5 ENST00000455537.7 ENST00000676457.1 |
KIF5A
|
kinesin family member 5A |
| chr1_-_109397888 | 0.09 |
ENST00000256637.8
|
SORT1
|
sortilin 1 |
| chrX_+_16946650 | 0.09 |
ENST00000357277.8
|
REPS2
|
RALBP1 associated Eps domain containing 2 |
| chr1_+_109249530 | 0.09 |
ENST00000271332.4
|
CELSR2
|
cadherin EGF LAG seven-pass G-type receptor 2 |
| chr21_-_38498415 | 0.09 |
ENST00000398905.5
ENST00000398907.5 ENST00000453032.6 ENST00000288319.12 |
ERG
|
ETS transcription factor ERG |
| chr14_+_105314711 | 0.09 |
ENST00000447393.6
ENST00000547217.5 |
PACS2
|
phosphofurin acidic cluster sorting protein 2 |
| chr17_-_45490696 | 0.09 |
ENST00000430334.8
ENST00000584420.1 ENST00000589780.5 |
PLEKHM1
|
pleckstrin homology and RUN domain containing M1 |
| chr10_+_96043394 | 0.09 |
ENST00000403870.7
ENST00000265992.9 ENST00000465148.3 |
CCNJ
|
cyclin J |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:2000974 | trochlear nerve development(GO:0021558) auditory receptor cell fate determination(GO:0042668) negative regulation of auditory receptor cell differentiation(GO:0045608) regulation of timing of neuron differentiation(GO:0060164) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.2 | 0.5 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.1 | 0.3 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.1 | 0.5 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.1 | 0.4 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.1 | 0.9 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.1 | 0.2 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 0.5 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) |
| 0.1 | 0.5 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.1 | 0.4 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.1 | 0.3 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.5 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 | 0.2 | GO:0060268 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of respiratory burst(GO:0060268) |
| 0.1 | 0.2 | GO:1900276 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.2 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0061445 | endocardial cell fate commitment(GO:0060957) endocardial cushion cell fate commitment(GO:0061445) |
| 0.0 | 0.2 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.8 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.0 | 0.6 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.1 | GO:0061386 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.3 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.2 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.1 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.1 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.2 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.1 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.4 | GO:0032230 | positive regulation of synaptic transmission, GABAergic(GO:0032230) |
| 0.0 | 0.3 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.1 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.1 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 0.3 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.0 | 0.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.0 | 0.9 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 0.1 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.3 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.0 | GO:0072106 | regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.0 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.1 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.1 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.0 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.0 | 0.1 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.0 | 0.1 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.1 | GO:0061528 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.0 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.2 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.2 | GO:0042415 | norepinephrine metabolic process(GO:0042415) surfactant homeostasis(GO:0043129) myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.0 | GO:1903061 | positive regulation of protein lipidation(GO:1903061) |
| 0.0 | 0.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.4 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.0 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 0.0 | 0.1 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.1 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.0 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.2 | GO:0060746 | maternal behavior(GO:0042711) parental behavior(GO:0060746) |
| 0.0 | 0.0 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.0 | 0.1 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:0098736 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) negative regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901205) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.2 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.0 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0061428 | peptidyl-aspartic acid hydroxylation(GO:0042264) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.3 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.0 | 0.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.1 | 0.3 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 0.4 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 0.2 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 0.2 | GO:0039713 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.3 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0030849 | X chromosome(GO:0000805) autosome(GO:0030849) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.2 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.1 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.2 | 0.5 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.1 | 0.4 | GO:0016524 | latrotoxin receptor activity(GO:0016524) |
| 0.1 | 0.4 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.1 | 0.2 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.1 | 0.9 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.2 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.4 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.3 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.3 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.2 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.5 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.0 | 0.3 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.1 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.1 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.1 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.0 | 0.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.0 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.0 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.0 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.6 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |