Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for VAX2_RHOXF2
Z-value: 0.41
Transcription factors associated with VAX2_RHOXF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
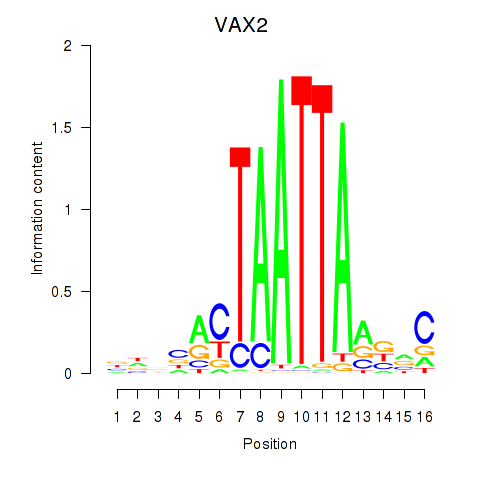
VAX2
|
ENSG00000116035.4 | VAX2 |
|
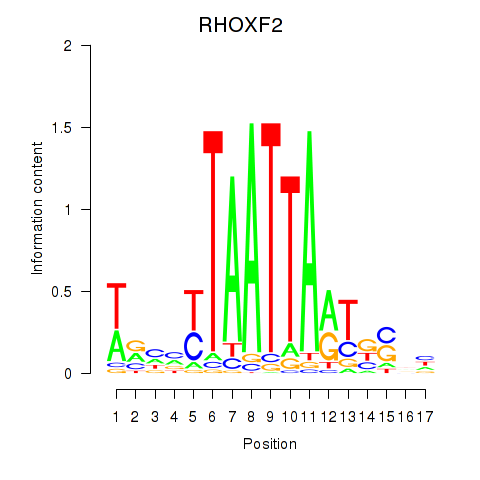
RHOXF2
|
ENSG00000131721.6 | RHOXF2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RHOXF2 | hg38_v1_chrX_+_120158600_120158624 | -0.14 | 4.9e-01 | Click! |
| VAX2 | hg38_v1_chr2_+_70900546_70900617 | 0.06 | 7.9e-01 | Click! |
Activity profile of VAX2_RHOXF2 motif
Sorted Z-values of VAX2_RHOXF2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of VAX2_RHOXF2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_26195647 | 1.05 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr6_+_26402237 | 0.76 |
ENST00000476549.6
ENST00000450085.6 ENST00000425234.6 ENST00000427334.5 ENST00000506698.1 ENST00000289361.11 |
BTN3A1
|
butyrophilin subfamily 3 member A1 |
| chr6_+_26440472 | 0.68 |
ENST00000494393.5
ENST00000482451.5 ENST00000471353.5 ENST00000361232.7 ENST00000487627.5 ENST00000496719.1 ENST00000244519.7 ENST00000490254.5 ENST00000487272.1 |
BTN3A3
|
butyrophilin subfamily 3 member A3 |
| chr3_-_191282383 | 0.64 |
ENST00000427544.6
|
UTS2B
|
urotensin 2B |
| chr6_+_26365159 | 0.51 |
ENST00000532865.5
ENST00000396934.7 ENST00000508906.6 ENST00000530653.5 ENST00000527417.5 |
BTN3A2
|
butyrophilin subfamily 3 member A2 |
| chr11_-_117877463 | 0.50 |
ENST00000527717.5
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr11_-_117876719 | 0.47 |
ENST00000529335.6
ENST00000260282.8 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr11_-_117876892 | 0.45 |
ENST00000539526.5
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr12_+_26195313 | 0.44 |
ENST00000422622.3
|
SSPN
|
sarcospan |
| chr6_+_26365176 | 0.44 |
ENST00000377708.7
|
BTN3A2
|
butyrophilin subfamily 3 member A2 |
| chr1_-_56579555 | 0.43 |
ENST00000371250.4
|
PLPP3
|
phospholipid phosphatase 3 |
| chr6_+_26365215 | 0.42 |
ENST00000527422.5
ENST00000356386.6 ENST00000396948.5 |
BTN3A2
|
butyrophilin subfamily 3 member A2 |
| chr21_-_36542600 | 0.41 |
ENST00000399136.5
|
CLDN14
|
claudin 14 |
| chr17_-_42185452 | 0.40 |
ENST00000293330.1
|
HCRT
|
hypocretin neuropeptide precursor |
| chr12_+_26195543 | 0.39 |
ENST00000242729.7
|
SSPN
|
sarcospan |
| chr5_+_132873660 | 0.26 |
ENST00000296877.3
|
LEAP2
|
liver enriched antimicrobial peptide 2 |
| chr15_-_89211803 | 0.25 |
ENST00000563254.1
|
RLBP1
|
retinaldehyde binding protein 1 |
| chr7_+_138460238 | 0.24 |
ENST00000343526.9
|
TRIM24
|
tripartite motif containing 24 |
| chr19_-_3557563 | 0.23 |
ENST00000389395.7
ENST00000355415.7 |
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr18_+_58341038 | 0.22 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr9_+_122370523 | 0.21 |
ENST00000643810.1
ENST00000540753.6 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr2_+_89884740 | 0.20 |
ENST00000509129.1
|
IGKV1D-37
|
immunoglobulin kappa variable 1D-37 (non-functional) |
| chr2_-_89297785 | 0.20 |
ENST00000465170.1
|
IGKV1-37
|
immunoglobulin kappa variable 1-37 (non-functional) |
| chr3_-_142029108 | 0.19 |
ENST00000497579.5
|
TFDP2
|
transcription factor Dp-2 |
| chr5_-_20575850 | 0.19 |
ENST00000507958.5
|
CDH18
|
cadherin 18 |
| chr6_+_26402289 | 0.19 |
ENST00000414912.2
|
BTN3A1
|
butyrophilin subfamily 3 member A1 |
| chr11_+_72189528 | 0.19 |
ENST00000312293.9
|
FOLR1
|
folate receptor alpha |
| chr11_+_72189659 | 0.18 |
ENST00000393681.6
|
FOLR1
|
folate receptor alpha |
| chr20_-_31390483 | 0.18 |
ENST00000376315.2
|
DEFB119
|
defensin beta 119 |
| chr12_+_130953898 | 0.17 |
ENST00000261654.10
|
ADGRD1
|
adhesion G protein-coupled receptor D1 |
| chr3_+_178419123 | 0.16 |
ENST00000614557.1
ENST00000455307.5 ENST00000436432.1 |
ENSG00000275163.1
LINC01014
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 (KCNMB2-IT1 - KCNMB2 readthrough transcript) long intergenic non-protein coding RNA 1014 |
| chr4_+_94974984 | 0.16 |
ENST00000672698.1
|
BMPR1B
|
bone morphogenetic protein receptor type 1B |
| chr16_-_28597042 | 0.16 |
ENST00000533150.5
ENST00000335715.9 |
SULT1A2
|
sulfotransferase family 1A member 2 |
| chr11_+_24496988 | 0.15 |
ENST00000336930.11
|
LUZP2
|
leucine zipper protein 2 |
| chr11_-_35419098 | 0.15 |
ENST00000606205.6
ENST00000645303.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr11_-_35419213 | 0.14 |
ENST00000642171.1
ENST00000644050.1 ENST00000643134.1 |
SLC1A2
|
solute carrier family 1 member 2 |
| chr4_+_87832917 | 0.14 |
ENST00000395102.8
ENST00000497649.6 ENST00000540395.1 ENST00000560249.5 ENST00000511670.5 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr8_-_100649660 | 0.14 |
ENST00000311812.7
|
SNX31
|
sorting nexin 31 |
| chr4_+_87650277 | 0.13 |
ENST00000339673.11
ENST00000282479.8 |
DMP1
|
dentin matrix acidic phosphoprotein 1 |
| chr1_+_159204860 | 0.13 |
ENST00000368122.4
ENST00000368121.6 |
ACKR1
|
atypical chemokine receptor 1 (Duffy blood group) |
| chr14_+_32329256 | 0.12 |
ENST00000280979.9
|
AKAP6
|
A-kinase anchoring protein 6 |
| chr5_+_129748091 | 0.11 |
ENST00000564719.2
|
MINAR2
|
membrane integral NOTCH2 associated receptor 2 |
| chr20_+_33217325 | 0.11 |
ENST00000375452.3
ENST00000375454.8 |
BPIFA3
|
BPI fold containing family A member 3 |
| chr4_+_87975667 | 0.11 |
ENST00000237623.11
ENST00000682655.1 ENST00000508233.6 ENST00000360804.4 ENST00000395080.8 |
SPP1
|
secreted phosphoprotein 1 |
| chr20_+_57561103 | 0.11 |
ENST00000319441.6
|
PCK1
|
phosphoenolpyruvate carboxykinase 1 |
| chr3_+_157436842 | 0.11 |
ENST00000295927.4
|
PTX3
|
pentraxin 3 |
| chr14_+_103385450 | 0.11 |
ENST00000416682.6
|
MARK3
|
microtubule affinity regulating kinase 3 |
| chr13_+_108629605 | 0.11 |
ENST00000457511.7
|
MYO16
|
myosin XVI |
| chr15_+_58138368 | 0.10 |
ENST00000219919.9
ENST00000536493.1 |
AQP9
|
aquaporin 9 |
| chr2_+_90220727 | 0.10 |
ENST00000471857.2
|
IGKV1D-8
|
immunoglobulin kappa variable 1D-8 |
| chr4_+_87975829 | 0.10 |
ENST00000614857.5
|
SPP1
|
secreted phosphoprotein 1 |
| chr12_+_80716906 | 0.10 |
ENST00000228644.4
|
MYF5
|
myogenic factor 5 |
| chr14_-_52791597 | 0.10 |
ENST00000216410.8
ENST00000557604.1 |
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr14_+_32329341 | 0.10 |
ENST00000557354.5
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A-kinase anchoring protein 6 |
| chr4_+_75556048 | 0.10 |
ENST00000616557.1
ENST00000435974.2 ENST00000311623.9 |
ODAPH
|
odontogenesis associated phosphoprotein |
| chr8_+_91249307 | 0.09 |
ENST00000309536.6
ENST00000276609.8 |
SLC26A7
|
solute carrier family 26 member 7 |
| chr1_-_92486916 | 0.09 |
ENST00000294702.6
|
GFI1
|
growth factor independent 1 transcriptional repressor |
| chr7_-_44541318 | 0.09 |
ENST00000381160.8
|
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr11_+_33039996 | 0.09 |
ENST00000432887.5
ENST00000528898.1 ENST00000531632.6 |
TCP11L1
|
t-complex 11 like 1 |
| chr10_+_18400562 | 0.09 |
ENST00000377315.5
ENST00000650685.1 |
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr10_+_18260715 | 0.09 |
ENST00000615785.4
ENST00000617363.4 ENST00000396576.6 |
CACNB2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr2_-_74392025 | 0.09 |
ENST00000440727.1
ENST00000409240.5 |
DCTN1
|
dynactin subunit 1 |
| chr14_-_55191534 | 0.09 |
ENST00000395425.6
ENST00000247191.7 |
DLGAP5
|
DLG associated protein 5 |
| chr9_-_134068012 | 0.09 |
ENST00000303407.12
|
BRD3
|
bromodomain containing 3 |
| chr2_-_189179754 | 0.09 |
ENST00000374866.9
ENST00000618828.1 |
COL5A2
|
collagen type V alpha 2 chain |
| chr19_-_14979848 | 0.08 |
ENST00000594383.2
|
SLC1A6
|
solute carrier family 1 member 6 |
| chr9_+_122371036 | 0.08 |
ENST00000619306.5
ENST00000426608.6 ENST00000223423.8 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr5_+_67004618 | 0.08 |
ENST00000261569.11
ENST00000436277.5 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr6_+_29550407 | 0.08 |
ENST00000641137.1
|
OR2I1P
|
olfactory receptor family 2 subfamily I member 1 pseudogene |
| chr1_-_211134135 | 0.08 |
ENST00000638983.1
ENST00000271751.10 ENST00000639952.1 |
ENSG00000284299.1
KCNH1
|
novel protein potassium voltage-gated channel subfamily H member 1 |
| chr3_+_42979281 | 0.08 |
ENST00000488863.5
ENST00000430121.3 |
GASK1A
|
golgi associated kinase 1A |
| chr8_-_33567118 | 0.08 |
ENST00000256257.2
|
RNF122
|
ring finger protein 122 |
| chr10_+_70052582 | 0.08 |
ENST00000676699.1
|
MACROH2A2
|
macroH2A.2 histone |
| chr17_-_41047267 | 0.07 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr5_+_172641241 | 0.07 |
ENST00000369800.6
ENST00000520919.5 ENST00000522853.5 |
NEURL1B
|
neuralized E3 ubiquitin protein ligase 1B |
| chr6_-_82247697 | 0.07 |
ENST00000306270.12
ENST00000610980.4 |
IBTK
|
inhibitor of Bruton tyrosine kinase |
| chrX_+_73563190 | 0.07 |
ENST00000373504.10
ENST00000373502.9 |
CHIC1
|
cysteine rich hydrophobic domain 1 |
| chr7_-_44082464 | 0.07 |
ENST00000335195.10
ENST00000395831.7 ENST00000414235.5 ENST00000242248.10 ENST00000452049.1 |
POLM
|
DNA polymerase mu |
| chrX_+_136648138 | 0.07 |
ENST00000370629.7
|
CD40LG
|
CD40 ligand |
| chr11_+_124183219 | 0.07 |
ENST00000641351.2
|
OR10D3
|
olfactory receptor family 10 subfamily D member 3 |
| chr17_-_10469558 | 0.06 |
ENST00000255381.2
|
MYH4
|
myosin heavy chain 4 |
| chr1_-_211134061 | 0.06 |
ENST00000639602.1
ENST00000638498.1 ENST00000367007.5 |
ENSG00000284299.1
KCNH1
|
novel protein potassium voltage-gated channel subfamily H member 1 |
| chr14_+_103385374 | 0.06 |
ENST00000678179.1
ENST00000676938.1 ENST00000678619.1 ENST00000440884.7 ENST00000560417.6 ENST00000679330.1 ENST00000556744.2 ENST00000676897.1 ENST00000677560.1 ENST00000561314.6 ENST00000677829.1 ENST00000677133.1 ENST00000676645.1 ENST00000678175.1 ENST00000429436.7 ENST00000677360.1 ENST00000678237.1 ENST00000677347.1 ENST00000677432.1 |
MARK3
|
microtubule affinity regulating kinase 3 |
| chr2_-_74391837 | 0.06 |
ENST00000417090.1
ENST00000409868.5 ENST00000680606.1 |
DCTN1
|
dynactin subunit 1 |
| chr16_+_69311339 | 0.06 |
ENST00000254950.13
|
VPS4A
|
vacuolar protein sorting 4 homolog A |
| chr19_+_15728024 | 0.06 |
ENST00000305899.5
|
OR10H2
|
olfactory receptor family 10 subfamily H member 2 |
| chr7_+_71132123 | 0.06 |
ENST00000333538.10
|
GALNT17
|
polypeptide N-acetylgalactosaminyltransferase 17 |
| chr2_-_213151590 | 0.06 |
ENST00000374319.8
ENST00000457361.5 ENST00000451136.6 ENST00000434687.6 |
IKZF2
|
IKAROS family zinc finger 2 |
| chr14_+_64549950 | 0.06 |
ENST00000298705.6
|
PPP1R36
|
protein phosphatase 1 regulatory subunit 36 |
| chr1_-_152414256 | 0.06 |
ENST00000271835.3
|
CRNN
|
cornulin |
| chr18_+_34976928 | 0.06 |
ENST00000591734.5
ENST00000413393.5 ENST00000589180.5 ENST00000587359.5 |
MAPRE2
|
microtubule associated protein RP/EB family member 2 |
| chr4_+_168497044 | 0.05 |
ENST00000505667.6
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr15_-_49620747 | 0.05 |
ENST00000559905.5
ENST00000560246.1 ENST00000299338.11 ENST00000558594.5 |
FAM227B
|
family with sequence similarity 227 member B |
| chr8_-_96261579 | 0.05 |
ENST00000517720.1
ENST00000523821.5 ENST00000287025.4 |
MTERF3
|
mitochondrial transcription termination factor 3 |
| chr1_+_174700413 | 0.05 |
ENST00000529145.6
ENST00000325589.9 |
RABGAP1L
|
RAB GTPase activating protein 1 like |
| chr9_+_12693327 | 0.05 |
ENST00000388918.10
|
TYRP1
|
tyrosinase related protein 1 |
| chrX_+_136648214 | 0.05 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr8_-_39838250 | 0.05 |
ENST00000622267.1
|
ADAM2
|
ADAM metallopeptidase domain 2 |
| chr4_+_168497066 | 0.04 |
ENST00000261509.10
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr1_-_113871665 | 0.04 |
ENST00000528414.5
ENST00000460620.5 ENST00000359785.10 ENST00000420377.6 ENST00000525799.1 ENST00000538253.5 |
PTPN22
|
protein tyrosine phosphatase non-receptor type 22 |
| chr18_-_69956924 | 0.04 |
ENST00000581982.5
ENST00000280200.8 |
CD226
|
CD226 molecule |
| chr6_+_54018910 | 0.04 |
ENST00000514921.5
ENST00000274897.9 ENST00000370877.6 |
MLIP
|
muscular LMNA interacting protein |
| chr19_-_45584769 | 0.04 |
ENST00000263275.5
|
OPA3
|
outer mitochondrial membrane lipid metabolism regulator OPA3 |
| chr6_+_29099490 | 0.04 |
ENST00000641659.2
|
OR2J1
|
olfactory receptor family 2 subfamily J member 1 |
| chr20_+_15196834 | 0.04 |
ENST00000402914.5
|
MACROD2
|
mono-ADP ribosylhydrolase 2 |
| chr20_-_52105644 | 0.04 |
ENST00000371523.8
|
ZFP64
|
ZFP64 zinc finger protein |
| chr4_-_159035226 | 0.04 |
ENST00000434826.3
|
C4orf45
|
chromosome 4 open reading frame 45 |
| chr4_-_145180496 | 0.04 |
ENST00000447906.8
|
OTUD4
|
OTU deubiquitinase 4 |
| chr12_-_118359639 | 0.04 |
ENST00000541786.5
ENST00000419821.6 ENST00000541878.5 |
TAOK3
|
TAO kinase 3 |
| chr5_+_177384430 | 0.04 |
ENST00000512593.5
ENST00000324417.6 |
SLC34A1
|
solute carrier family 34 member 1 |
| chr3_-_151316795 | 0.03 |
ENST00000260843.5
|
GPR87
|
G protein-coupled receptor 87 |
| chr5_+_121961955 | 0.03 |
ENST00000339397.5
|
SRFBP1
|
serum response factor binding protein 1 |
| chr8_-_39838201 | 0.03 |
ENST00000347580.8
ENST00000379853.6 ENST00000265708.9 ENST00000521880.5 |
ADAM2
|
ADAM metallopeptidase domain 2 |
| chr15_+_49170237 | 0.03 |
ENST00000560031.6
ENST00000558145.5 ENST00000544523.5 ENST00000560138.5 |
GALK2
|
galactokinase 2 |
| chr16_+_7303245 | 0.03 |
ENST00000674626.1
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr11_-_7941708 | 0.02 |
ENST00000642047.1
|
OR10A3
|
olfactory receptor family 10 subfamily A member 3 |
| chr20_-_51802509 | 0.02 |
ENST00000371539.7
ENST00000217086.9 |
SALL4
|
spalt like transcription factor 4 |
| chr4_+_150582119 | 0.02 |
ENST00000317605.6
|
MAB21L2
|
mab-21 like 2 |
| chr14_+_71933116 | 0.02 |
ENST00000553530.5
ENST00000556437.5 |
RGS6
|
regulator of G protein signaling 6 |
| chr8_-_90082871 | 0.02 |
ENST00000265431.7
|
CALB1
|
calbindin 1 |
| chr1_+_75134382 | 0.02 |
ENST00000356261.4
|
LHX8
|
LIM homeobox 8 |
| chr7_-_44541262 | 0.02 |
ENST00000289547.8
ENST00000546276.5 ENST00000423141.1 |
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr10_-_97445850 | 0.02 |
ENST00000477692.6
ENST00000485122.6 ENST00000370886.9 ENST00000370885.8 ENST00000370902.8 ENST00000370884.5 |
EXOSC1
|
exosome component 1 |
| chr10_+_97446137 | 0.02 |
ENST00000370854.7
ENST00000393760.6 ENST00000414567.5 |
ZDHHC16
|
zinc finger DHHC-type palmitoyltransferase 16 |
| chr5_+_90474879 | 0.02 |
ENST00000504930.5
ENST00000514483.5 |
POLR3G
|
RNA polymerase III subunit G |
| chr7_+_73830988 | 0.02 |
ENST00000340958.4
|
CLDN4
|
claudin 4 |
| chr11_-_111923722 | 0.02 |
ENST00000527950.5
|
CRYAB
|
crystallin alpha B |
| chr10_+_72893734 | 0.01 |
ENST00000334011.10
|
OIT3
|
oncoprotein induced transcript 3 |
| chr19_+_41262480 | 0.01 |
ENST00000352456.7
ENST00000595018.5 ENST00000597725.5 |
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U like 1 |
| chr5_+_90474848 | 0.01 |
ENST00000651687.1
|
POLR3G
|
RNA polymerase III subunit G |
| chr17_+_45161070 | 0.01 |
ENST00000593138.6
ENST00000586681.6 |
HEXIM2
|
HEXIM P-TEFb complex subunit 2 |
| chr10_+_97446194 | 0.01 |
ENST00000370846.8
ENST00000352634.8 ENST00000353979.7 ENST00000370842.6 ENST00000345745.9 |
ZDHHC16
|
zinc finger DHHC-type palmitoyltransferase 16 |
| chr9_-_34895722 | 0.01 |
ENST00000603640.6
ENST00000603592.1 ENST00000340783.11 |
FAM205C
|
family with sequence similarity 205 member C |
| chr11_-_107858777 | 0.01 |
ENST00000525815.6
|
SLC35F2
|
solute carrier family 35 member F2 |
| chr4_-_73620629 | 0.01 |
ENST00000342081.7
|
RASSF6
|
Ras association domain family member 6 |
| chr7_-_111392915 | 0.01 |
ENST00000450877.5
|
IMMP2L
|
inner mitochondrial membrane peptidase subunit 2 |
| chr20_-_51802433 | 0.01 |
ENST00000395997.3
|
SALL4
|
spalt like transcription factor 4 |
| chr17_+_7407838 | 0.01 |
ENST00000302926.7
|
NLGN2
|
neuroligin 2 |
| chr12_-_10802627 | 0.01 |
ENST00000240687.2
|
TAS2R7
|
taste 2 receptor member 7 |
| chr14_+_56117702 | 0.01 |
ENST00000559044.5
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr7_-_101217569 | 0.01 |
ENST00000223127.8
|
PLOD3
|
procollagen-lysine,2-oxoglutarate 5-dioxygenase 3 |
| chr11_-_5301946 | 0.00 |
ENST00000380224.2
|
OR51B4
|
olfactory receptor family 51 subfamily B member 4 |
| chr17_-_40665121 | 0.00 |
ENST00000394052.5
|
KRT222
|
keratin 222 |
| chr17_+_50746534 | 0.00 |
ENST00000511974.5
|
LUC7L3
|
LUC7 like 3 pre-mRNA splicing factor |
| chr7_-_100119840 | 0.00 |
ENST00000437822.6
|
TAF6
|
TATA-box binding protein associated factor 6 |
| chr6_+_106360668 | 0.00 |
ENST00000633556.3
|
CRYBG1
|
crystallin beta-gamma domain containing 1 |
| chrX_-_155071064 | 0.00 |
ENST00000369484.8
ENST00000369476.8 |
CMC4
MTCP1
|
C-X9-C motif containing 4 mature T cell proliferation 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.1 | 2.3 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.1 | 0.4 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.2 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.0 | 0.1 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 0.0 | 0.1 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.4 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0015855 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.2 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.0 | 0.4 | GO:0098712 | L-glutamate import across plasma membrane(GO:0098712) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.2 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.2 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 1.4 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.0 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.0 | GO:0097187 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 1.9 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.1 | 0.4 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.2 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 1.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |