Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
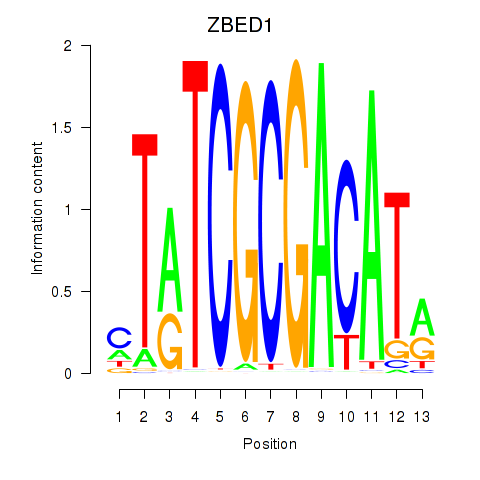
Results for ZBED1
Z-value: 0.29
Transcription factors associated with ZBED1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBED1
|
ENSG00000214717.12 | ZBED1 |
|
ZBED1
|
ENSG00000214717.12 | ZBED1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBED1 | hg38_v1_chrX_-_2500895_2500983, hg38_v1_chrY_-_2500895_2500976 | -0.49 | 1.3e-02 | Click! |
Activity profile of ZBED1 motif
Sorted Z-values of ZBED1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBED1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.1 | 0.4 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.5 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0031417 | NatC complex(GO:0031417) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |