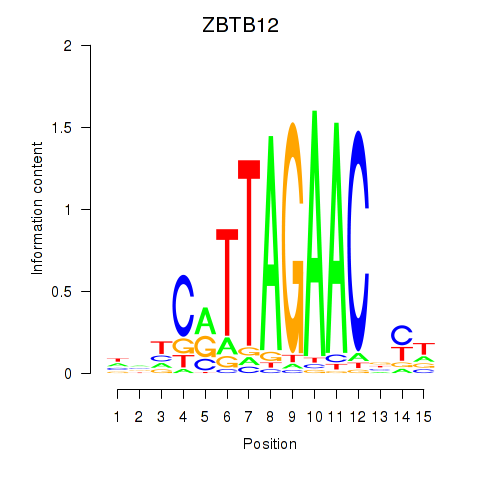
|
chr1_+_61404076
Show fit
|
2.44 |
ENST00000357977.5
|
NFIA
|
nuclear factor I A
|
|
chr7_+_90154442
Show fit
|
1.34 |
ENST00000297205.7
|
STEAP1
|
STEAP family member 1
|
|
chr5_-_88883701
Show fit
|
1.01 |
ENST00000636998.1
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr5_-_88883147
Show fit
|
0.94 |
ENST00000513252.5
ENST00000506554.5
ENST00000508569.5
ENST00000637732.1
ENST00000504921.7
ENST00000637481.1
ENST00000510942.5
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr3_-_149333619
Show fit
|
0.91 |
ENST00000296059.7
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr3_-_149333407
Show fit
|
0.89 |
ENST00000470080.5
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr14_-_20802836
Show fit
|
0.84 |
ENST00000397967.5
ENST00000555698.5
ENST00000397970.4
ENST00000340900.3
|
RNASE1
|
ribonuclease A family member 1, pancreatic
|
|
chr5_-_38557459
Show fit
|
0.73 |
ENST00000511561.1
|
LIFR
|
LIF receptor subunit alpha
|
|
chr5_-_88883420
Show fit
|
0.69 |
ENST00000437473.6
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr8_+_2045037
Show fit
|
0.68 |
ENST00000262113.9
|
MYOM2
|
myomesin 2
|
|
chr17_+_4740005
Show fit
|
0.64 |
ENST00000269289.10
|
ZMYND15
|
zinc finger MYND-type containing 15
|
|
chr12_-_46372763
Show fit
|
0.63 |
ENST00000256689.10
|
SLC38A2
|
solute carrier family 38 member 2
|
|
chr5_-_88883199
Show fit
|
0.61 |
ENST00000514015.5
ENST00000503075.1
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr8_+_2045058
Show fit
|
0.61 |
ENST00000523438.1
|
MYOM2
|
myomesin 2
|
|
chr2_-_105396943
Show fit
|
0.60 |
ENST00000409807.5
|
FHL2
|
four and a half LIM domains 2
|
|
chr17_+_4740042
Show fit
|
0.60 |
ENST00000592813.5
|
ZMYND15
|
zinc finger MYND-type containing 15
|
|
chr1_+_45583846
Show fit
|
0.58 |
ENST00000437901.6
ENST00000537798.5
ENST00000350030.8
ENST00000527470.5
ENST00000525515.5
ENST00000528238.5
ENST00000470768.5
ENST00000372052.8
ENST00000629893.1
ENST00000351223.7
|
NASP
|
nuclear autoantigenic sperm protein
|
|
chr6_+_2988606
Show fit
|
0.57 |
ENST00000380472.7
ENST00000605901.1
ENST00000454015.1
|
NQO2
LINC01011
|
N-ribosyldihydronicotinamide:quinone reductase 2
long intergenic non-protein coding RNA 1011
|
|
chr11_-_2149603
Show fit
|
0.56 |
ENST00000643349.1
|
ENSG00000284779.2
|
novel protein
|
|
chr22_+_50089879
Show fit
|
0.56 |
ENST00000545383.5
ENST00000262794.10
|
MOV10L1
|
Mov10 like RISC complex RNA helicase 1
|
|
chr2_+_189857393
Show fit
|
0.55 |
ENST00000452382.1
|
PMS1
|
PMS1 homolog 1, mismatch repair system component
|
|
chr1_+_159009886
Show fit
|
0.54 |
ENST00000340979.10
ENST00000368131.8
ENST00000295809.12
ENST00000368132.7
|
IFI16
|
interferon gamma inducible protein 16
|
|
chr9_+_124853417
Show fit
|
0.53 |
ENST00000613760.4
ENST00000618744.4
ENST00000373574.2
|
WDR38
|
WD repeat domain 38
|
|
chr1_+_159010002
Show fit
|
0.52 |
ENST00000359709.7
|
IFI16
|
interferon gamma inducible protein 16
|
|
chr22_+_50090028
Show fit
|
0.50 |
ENST00000395858.7
|
MOV10L1
|
Mov10 like RISC complex RNA helicase 1
|
|
chr6_+_18387326
Show fit
|
0.50 |
ENST00000259939.4
|
RNF144B
|
ring finger protein 144B
|
|
chr18_-_55586092
Show fit
|
0.49 |
ENST00000563888.6
ENST00000540999.5
ENST00000627685.2
|
TCF4
|
transcription factor 4
|
|
chr10_+_5412542
Show fit
|
0.48 |
ENST00000355029.9
|
NET1
|
neuroepithelial cell transforming 1
|
|
chr19_+_39296399
Show fit
|
0.48 |
ENST00000333625.3
|
IFNL1
|
interferon lambda 1
|
|
chr19_+_54906140
Show fit
|
0.47 |
ENST00000291890.9
ENST00000598576.5
ENST00000594765.5
ENST00000350790.9
ENST00000338835.9
ENST00000357397.5
|
NCR1
|
natural cytotoxicity triggering receptor 1
|
|
chr10_+_129467178
Show fit
|
0.47 |
ENST00000306010.8
ENST00000651593.1
|
MGMT
|
O-6-methylguanine-DNA methyltransferase
|
|
chr18_-_55510753
Show fit
|
0.45 |
ENST00000543082.5
|
TCF4
|
transcription factor 4
|
|
chr18_-_55423757
Show fit
|
0.45 |
ENST00000675707.1
|
TCF4
|
transcription factor 4
|
|
chr13_-_77919390
Show fit
|
0.42 |
ENST00000475537.2
ENST00000646605.1
|
EDNRB
|
endothelin receptor type B
|
|
chr1_+_13585453
Show fit
|
0.42 |
ENST00000487038.5
ENST00000475043.5
|
PDPN
|
podoplanin
|
|
chr20_-_25339731
Show fit
|
0.42 |
ENST00000450393.5
ENST00000491682.5
|
ABHD12
|
abhydrolase domain containing 12, lysophospholipase
|
|
chr7_+_142492121
Show fit
|
0.40 |
ENST00000390374.3
|
TRBV7-6
|
T cell receptor beta variable 7-6
|
|
chr5_+_42548043
Show fit
|
0.40 |
ENST00000618088.4
ENST00000612382.4
|
GHR
|
growth hormone receptor
|
|
chr17_-_41521719
Show fit
|
0.39 |
ENST00000393976.6
|
KRT15
|
keratin 15
|
|
chr20_-_38165261
Show fit
|
0.37 |
ENST00000361475.7
|
TGM2
|
transglutaminase 2
|
|
chr20_-_63463843
Show fit
|
0.36 |
ENST00000637193.1
|
KCNQ2
|
potassium voltage-gated channel subfamily Q member 2
|
|
chrX_-_134797134
Show fit
|
0.35 |
ENST00000370790.5
ENST00000493333.5
ENST00000611027.2
ENST00000343004.10
ENST00000298090.10
|
PABIR2
|
PABIR family member 2
|
|
chr14_-_93955577
Show fit
|
0.34 |
ENST00000555507.5
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr11_+_123590939
Show fit
|
0.33 |
ENST00000646146.1
|
GRAMD1B
|
GRAM domain containing 1B
|
|
chrX_+_71118515
Show fit
|
0.32 |
ENST00000333646.10
|
MED12
|
mediator complex subunit 12
|
|
chr2_-_111884117
Show fit
|
0.32 |
ENST00000341068.8
|
ANAPC1
|
anaphase promoting complex subunit 1
|
|
chr14_-_54441325
Show fit
|
0.32 |
ENST00000556113.1
ENST00000553660.5
ENST00000216416.9
ENST00000395573.8
ENST00000557690.5
|
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1
|
|
chrX_+_71118576
Show fit
|
0.31 |
ENST00000374080.8
|
MED12
|
mediator complex subunit 12
|
|
chrX_+_71118675
Show fit
|
0.30 |
ENST00000374102.5
|
MED12
|
mediator complex subunit 12
|
|
chr5_-_123036664
Show fit
|
0.30 |
ENST00000306442.5
|
PPIC
|
peptidylprolyl isomerase C
|
|
chr5_-_43313473
Show fit
|
0.30 |
ENST00000433297.2
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1
|
|
chr3_-_114624193
Show fit
|
0.29 |
ENST00000481632.5
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr5_-_43313403
Show fit
|
0.28 |
ENST00000325110.11
|
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1
|
|
chr8_+_117520832
Show fit
|
0.28 |
ENST00000522839.1
|
MED30
|
mediator complex subunit 30
|
|
chr17_-_4739866
Show fit
|
0.28 |
ENST00000574412.6
ENST00000293778.12
|
CXCL16
|
C-X-C motif chemokine ligand 16
|
|
chr17_+_75784798
Show fit
|
0.27 |
ENST00000589666.6
|
UNK
|
unk zinc finger
|
|
chr8_+_117520696
Show fit
|
0.26 |
ENST00000297347.7
|
MED30
|
mediator complex subunit 30
|
|
chr1_-_201171545
Show fit
|
0.25 |
ENST00000367333.6
|
TMEM9
|
transmembrane protein 9
|
|
chr4_-_99219230
Show fit
|
0.25 |
ENST00000394897.5
ENST00000508558.1
ENST00000394899.6
|
ADH6
|
alcohol dehydrogenase 6 (class V)
|
|
chr11_+_112175526
Show fit
|
0.24 |
ENST00000532612.5
ENST00000438022.5
|
BCO2
|
beta-carotene oxygenase 2
|
|
chrX_-_24672654
Show fit
|
0.24 |
ENST00000379145.5
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr11_-_60184633
Show fit
|
0.23 |
ENST00000529054.5
ENST00000530839.5
ENST00000426738.6
|
MS4A6A
|
membrane spanning 4-domains A6A
|
|
chr5_+_36606355
Show fit
|
0.23 |
ENST00000681909.1
ENST00000513903.5
ENST00000681795.1
ENST00000680125.1
ENST00000612708.5
ENST00000680232.1
ENST00000681776.1
ENST00000681926.1
ENST00000679958.1
ENST00000265113.9
ENST00000504121.5
ENST00000512374.1
ENST00000613445.5
ENST00000679983.1
|
SLC1A3
|
solute carrier family 1 member 3
|
|
chr20_+_11892493
Show fit
|
0.23 |
ENST00000422390.5
ENST00000618918.4
|
BTBD3
|
BTB domain containing 3
|
|
chr22_-_21225554
Show fit
|
0.22 |
ENST00000405188.8
|
GGT2
|
gamma-glutamyltransferase 2
|
|
chr8_-_27837765
Show fit
|
0.22 |
ENST00000521226.2
ENST00000301905.9
|
PBK
|
PDZ binding kinase
|
|
chr7_+_90469634
Show fit
|
0.21 |
ENST00000509356.2
|
PTTG1IP2
|
PTTG1IP family member 2
|
|
chr14_-_70359630
Show fit
|
0.21 |
ENST00000389912.7
|
COX16
|
cytochrome c oxidase assembly factor COX16
|
|
chr1_+_161153968
Show fit
|
0.19 |
ENST00000368003.6
|
UFC1
|
ubiquitin-fold modifier conjugating enzyme 1
|
|
chr11_+_112176364
Show fit
|
0.19 |
ENST00000526088.5
ENST00000532593.5
ENST00000531169.5
|
BCO2
|
beta-carotene oxygenase 2
|
|
chr14_-_22469673
Show fit
|
0.18 |
ENST00000535880.2
|
TRDV3
|
T cell receptor delta variable 3
|
|
chr2_+_112482133
Show fit
|
0.18 |
ENST00000233336.7
|
TTL
|
tubulin tyrosine ligase
|
|
chr11_+_124012997
Show fit
|
0.18 |
ENST00000641521.1
ENST00000641722.1
|
OR10G4
|
olfactory receptor family 10 subfamily G member 4
|
|
chr10_-_7619660
Show fit
|
0.18 |
ENST00000613909.4
|
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain 5
|
|
chr5_+_141208697
Show fit
|
0.18 |
ENST00000624949.1
ENST00000622978.1
ENST00000239450.4
|
PCDHB12
|
protocadherin beta 12
|
|
chr4_-_188109601
Show fit
|
0.17 |
ENST00000682553.1
|
TRIML2
|
tripartite motif family like 2
|
|
chr6_+_152697888
Show fit
|
0.17 |
ENST00000367245.5
ENST00000529453.1
|
MYCT1
|
MYC target 1
|
|
chr11_-_119381629
Show fit
|
0.17 |
ENST00000260187.7
ENST00000455332.6
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr11_+_63506073
Show fit
|
0.17 |
ENST00000255684.10
ENST00000394618.9
|
LGALS12
|
galectin 12
|
|
chr1_+_201786325
Show fit
|
0.16 |
ENST00000438083.1
|
NAV1
|
neuron navigator 1
|
|
chr11_-_48983826
Show fit
|
0.16 |
ENST00000649162.1
|
TRIM51GP
|
tripartite motif-containing 51G, pseudogene
|
|
chr8_-_30812867
Show fit
|
0.16 |
ENST00000518243.5
|
PPP2CB
|
protein phosphatase 2 catalytic subunit beta
|
|
chr19_-_35528221
Show fit
|
0.16 |
ENST00000588674.5
ENST00000452271.7
ENST00000518157.1
|
SBSN
|
suprabasin
|
|
chr11_+_63506046
Show fit
|
0.16 |
ENST00000674247.1
|
LGALS12
|
galectin 12
|
|
chr3_+_63819368
Show fit
|
0.16 |
ENST00000616659.1
|
C3orf49
|
chromosome 3 open reading frame 49
|
|
chr12_+_48727431
Show fit
|
0.16 |
ENST00000548380.6
|
TEX49
|
testis expressed 49
|
|
chr12_-_55927756
Show fit
|
0.16 |
ENST00000549939.1
|
PYM1
|
PYM homolog 1, exon junction complex associated factor
|
|
chr11_-_790062
Show fit
|
0.14 |
ENST00000330106.5
|
CEND1
|
cell cycle exit and neuronal differentiation 1
|
|
chr19_-_17405554
Show fit
|
0.14 |
ENST00000252593.7
|
BST2
|
bone marrow stromal cell antigen 2
|
|
chr22_-_41688859
Show fit
|
0.14 |
ENST00000401959.6
ENST00000648674.1
|
SNU13
|
small nuclear ribonucleoprotein 13
|
|
chr10_+_134465
Show fit
|
0.14 |
ENST00000439456.5
ENST00000397962.8
ENST00000309776.8
ENST00000397959.7
|
ZMYND11
|
zinc finger MYND-type containing 11
|
|
chr11_+_63506339
Show fit
|
0.13 |
ENST00000340246.10
|
LGALS12
|
galectin 12
|
|
chrX_-_30577759
Show fit
|
0.13 |
ENST00000378962.4
|
TASL
|
TLR adaptor interacting with endolysosomal SLC15A4
|
|
chr17_-_29078857
Show fit
|
0.13 |
ENST00000359450.6
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1
|
|
chr19_+_45692637
Show fit
|
0.13 |
ENST00000012049.10
ENST00000366382.8
|
QPCTL
|
glutaminyl-peptide cyclotransferase like
|
|
chr9_+_26947039
Show fit
|
0.13 |
ENST00000519968.5
ENST00000433700.5
|
IFT74
|
intraflagellar transport 74
|
|
chr14_-_21022095
Show fit
|
0.12 |
ENST00000635386.1
|
NDRG2
|
NDRG family member 2
|
|
chr12_-_102481744
Show fit
|
0.12 |
ENST00000644491.1
|
IGF1
|
insulin like growth factor 1
|
|
chr1_+_100038087
Show fit
|
0.12 |
ENST00000370152.8
|
MFSD14A
|
major facilitator superfamily domain containing 14A
|
|
chr7_+_130344837
Show fit
|
0.12 |
ENST00000485477.5
ENST00000431780.6
|
CPA5
|
carboxypeptidase A5
|
|
chr20_-_34955635
Show fit
|
0.12 |
ENST00000644793.1
ENST00000642498.1
ENST00000646735.1
ENST00000644608.1
ENST00000651619.1
|
GSS
|
glutathione synthetase
|
|
chr19_+_41262480
Show fit
|
0.12 |
ENST00000352456.7
ENST00000595018.5
ENST00000597725.5
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U like 1
|
|
chr1_-_209784521
Show fit
|
0.12 |
ENST00000294811.2
|
C1orf74
|
chromosome 1 open reading frame 74
|
|
chr11_-_60184415
Show fit
|
0.12 |
ENST00000532169.5
ENST00000534596.5
|
MS4A6A
|
membrane spanning 4-domains A6A
|
|
chr6_+_152697934
Show fit
|
0.11 |
ENST00000532295.1
|
MYCT1
|
MYC target 1
|
|
chr1_+_45551031
Show fit
|
0.11 |
ENST00000481885.5
ENST00000471651.1
|
AKR1A1
|
aldo-keto reductase family 1 member A1
|
|
chr9_-_7799752
Show fit
|
0.11 |
ENST00000358227.5
|
DMAC1
|
distal membrane arm assembly complex 1
|
|
chr8_-_140718661
Show fit
|
0.11 |
ENST00000430260.6
|
PTK2
|
protein tyrosine kinase 2
|
|
chr6_-_55875583
Show fit
|
0.11 |
ENST00000370830.4
|
BMP5
|
bone morphogenetic protein 5
|
|
chr12_-_122730828
Show fit
|
0.11 |
ENST00000432564.3
|
HCAR1
|
hydroxycarboxylic acid receptor 1
|
|
chr7_-_75486286
Show fit
|
0.11 |
ENST00000615331.5
|
POM121C
|
POM121 transmembrane nucleoporin C
|
|
chr22_+_39570753
Show fit
|
0.10 |
ENST00000407673.5
ENST00000401624.5
ENST00000404898.5
ENST00000402142.4
|
CACNA1I
|
calcium voltage-gated channel subunit alpha1 I
|
|
chr18_-_35290184
Show fit
|
0.10 |
ENST00000589178.5
ENST00000592278.1
ENST00000333206.10
ENST00000592211.1
ENST00000420878.7
ENST00000610712.1
ENST00000586922.2
|
ZSCAN30
ENSG00000268573.1
|
zinc finger and SCAN domain containing 30
novel transcript
|
|
chr19_-_45645560
Show fit
|
0.10 |
ENST00000587152.6
|
EML2
|
EMAP like 2
|
|
chr3_+_35641421
Show fit
|
0.09 |
ENST00000449196.5
|
ARPP21
|
cAMP regulated phosphoprotein 21
|
|
chr20_-_46684467
Show fit
|
0.09 |
ENST00000372121.5
|
SLC13A3
|
solute carrier family 13 member 3
|
|
chrX_-_71068384
Show fit
|
0.09 |
ENST00000276105.3
ENST00000622259.4
|
SNX12
|
sorting nexin 12
|
|
chr17_+_11598456
Show fit
|
0.09 |
ENST00000579828.5
ENST00000262442.9
ENST00000454412.6
|
DNAH9
|
dynein axonemal heavy chain 9
|
|
chr1_+_158931539
Show fit
|
0.09 |
ENST00000368140.6
ENST00000368138.7
ENST00000392254.6
ENST00000392252.7
ENST00000368135.4
|
PYHIN1
|
pyrin and HIN domain family member 1
|
|
chr10_-_96513911
Show fit
|
0.09 |
ENST00000357947.4
|
TLL2
|
tolloid like 2
|
|
chr16_+_89873571
Show fit
|
0.09 |
ENST00000263347.11
ENST00000263346.13
|
TCF25
|
transcription factor 25
|
|
chr1_+_22451843
Show fit
|
0.09 |
ENST00000375647.5
ENST00000404138.5
ENST00000374651.8
ENST00000400239.6
|
ZBTB40
|
zinc finger and BTB domain containing 40
|
|
chr7_+_130344810
Show fit
|
0.09 |
ENST00000497503.5
ENST00000463587.5
ENST00000461828.5
ENST00000474905.6
ENST00000494311.1
ENST00000466363.6
|
CPA5
|
carboxypeptidase A5
|
|
chr12_-_13095664
Show fit
|
0.08 |
ENST00000337630.10
ENST00000545699.1
|
GSG1
|
germ cell associated 1
|
|
chr2_-_196068812
Show fit
|
0.08 |
ENST00000410072.5
ENST00000312428.11
|
DNAH7
|
dynein axonemal heavy chain 7
|
|
chr20_+_34241390
Show fit
|
0.07 |
ENST00000374954.4
|
ASIP
|
agouti signaling protein
|
|
chr18_-_49460630
Show fit
|
0.07 |
ENST00000675505.1
ENST00000442713.6
ENST00000269445.10
|
DYM
|
dymeclin
|
|
chr3_+_49199477
Show fit
|
0.07 |
ENST00000452691.7
ENST00000366429.2
|
IHO1
|
interactor of HORMAD1 1
|
|
chr9_+_87882871
Show fit
|
0.07 |
ENST00000325643.6
|
SPATA31E1
|
SPATA31 subfamily E member 1
|
|
chr19_-_8321354
Show fit
|
0.06 |
ENST00000301457.3
|
NDUFA7
|
NADH:ubiquinone oxidoreductase subunit A7
|
|
chrX_+_134237047
Show fit
|
0.06 |
ENST00000370809.4
ENST00000517294.5
|
CCDC160
|
coiled-coil domain containing 160
|
|
chr11_+_8683201
Show fit
|
0.06 |
ENST00000526562.5
ENST00000525981.1
|
RPL27A
|
ribosomal protein L27a
|
|
chr14_+_61762405
Show fit
|
0.06 |
ENST00000216294.5
|
SNAPC1
|
small nuclear RNA activating complex polypeptide 1
|
|
chr11_+_12110569
Show fit
|
0.05 |
ENST00000683283.1
ENST00000256194.8
|
MICAL2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2
|
|
chr7_+_23598144
Show fit
|
0.05 |
ENST00000410069.1
|
CCDC126
|
coiled-coil domain containing 126
|
|
chr1_+_151036578
Show fit
|
0.05 |
ENST00000368931.8
ENST00000295294.11
|
BNIPL
|
BCL2 interacting protein like
|
|
chr1_+_84408230
Show fit
|
0.05 |
ENST00000370662.3
|
DNASE2B
|
deoxyribonuclease 2 beta
|
|
chr14_-_74493275
Show fit
|
0.05 |
ENST00000541064.5
|
NPC2
|
NPC intracellular cholesterol transporter 2
|
|
chr6_+_26158115
Show fit
|
0.05 |
ENST00000377777.5
ENST00000289316.2
|
H2BC5
|
H2B clustered histone 5
|
|
chr10_+_134703
Show fit
|
0.04 |
ENST00000509513.6
|
ZMYND11
|
zinc finger MYND-type containing 11
|
|
chr14_-_74493291
Show fit
|
0.04 |
ENST00000238633.6
ENST00000555619.6
ENST00000434013.6
|
NPC2
|
NPC intracellular cholesterol transporter 2
|
|
chr13_+_45120493
Show fit
|
0.04 |
ENST00000340473.8
|
GTF2F2
|
general transcription factor IIF subunit 2
|
|
chr5_+_69217721
Show fit
|
0.04 |
ENST00000256441.5
|
MRPS36
|
mitochondrial ribosomal protein S36
|
|
chr19_-_52139904
Show fit
|
0.04 |
ENST00000597013.5
ENST00000596290.5
ENST00000600228.6
|
ZNF616
|
zinc finger protein 616
|
|
chr14_-_74493322
Show fit
|
0.04 |
ENST00000553490.5
ENST00000557510.5
|
NPC2
|
NPC intracellular cholesterol transporter 2
|
|
chr7_+_48171451
Show fit
|
0.04 |
ENST00000435803.6
|
ABCA13
|
ATP binding cassette subfamily A member 13
|
|
chr3_+_89107649
Show fit
|
0.03 |
ENST00000452448.6
ENST00000494014.1
|
EPHA3
|
EPH receptor A3
|
|
chr17_+_75261864
Show fit
|
0.03 |
ENST00000245539.11
ENST00000579002.5
|
MRPS7
|
mitochondrial ribosomal protein S7
|
|
chr11_+_111255982
Show fit
|
0.03 |
ENST00000637637.1
|
C11orf53
|
chromosome 11 open reading frame 53
|
|
chr2_+_196713117
Show fit
|
0.03 |
ENST00000409270.5
|
CCDC150
|
coiled-coil domain containing 150
|
|
chr5_-_177351522
Show fit
|
0.03 |
ENST00000513877.1
ENST00000515209.5
ENST00000514458.5
ENST00000502560.5
ENST00000303127.12
|
LMAN2
|
lectin, mannose binding 2
|
|
chr6_-_33192454
Show fit
|
0.03 |
ENST00000395194.1
ENST00000341947.7
ENST00000374708.8
|
COL11A2
|
collagen type XI alpha 2 chain
|
|
chr5_+_141182369
Show fit
|
0.02 |
ENST00000609684.3
ENST00000625044.1
ENST00000623407.1
ENST00000623884.1
|
PCDHB16
ENSG00000279068.1
|
protocadherin beta 16
novel transcript
|
|
chr13_+_42138042
Show fit
|
0.02 |
ENST00000628433.2
ENST00000536612.3
|
DGKH
|
diacylglycerol kinase eta
|
|
chr12_-_55927865
Show fit
|
0.02 |
ENST00000454792.2
ENST00000408946.7
|
PYM1
|
PYM homolog 1, exon junction complex associated factor
|
|
chr5_+_119629552
Show fit
|
0.02 |
ENST00000613773.4
ENST00000620555.4
ENST00000515256.5
ENST00000509264.1
|
FAM170A
|
family with sequence similarity 170 member A
|
|
chr16_-_30123203
Show fit
|
0.02 |
ENST00000395202.5
ENST00000395199.7
ENST00000263025.9
ENST00000322266.9
|
MAPK3
|
mitogen-activated protein kinase 3
|
|
chr10_+_22928030
Show fit
|
0.02 |
ENST00000409983.7
ENST00000298032.10
ENST00000409049.7
|
ARMC3
|
armadillo repeat containing 3
|
|
chr10_+_22928010
Show fit
|
0.01 |
ENST00000376528.8
|
ARMC3
|
armadillo repeat containing 3
|
|
chr7_+_102748972
Show fit
|
0.01 |
ENST00000413034.3
ENST00000409231.7
ENST00000418198.5
|
FAM185A
|
family with sequence similarity 185 member A
|
|
chr8_-_63086031
Show fit
|
0.01 |
ENST00000260116.5
|
TTPA
|
alpha tocopherol transfer protein
|
|
chr1_+_159015665
Show fit
|
0.01 |
ENST00000567661.5
ENST00000474473.1
|
IFI16
|
interferon gamma inducible protein 16
|
|
chr11_+_65040895
Show fit
|
0.01 |
ENST00000531072.1
ENST00000398846.6
|
SAC3D1
|
SAC3 domain containing 1
|
|
chr11_+_60378475
Show fit
|
0.01 |
ENST00000358246.5
|
MS4A7
|
membrane spanning 4-domains A7
|
|
chr8_-_54022441
Show fit
|
0.01 |
ENST00000396401.7
ENST00000521604.7
ENST00000640382.1
ENST00000640041.1
|
TCEA1
|
transcription elongation factor A1
|
|
chr10_-_124450027
Show fit
|
0.00 |
ENST00000451024.5
|
NKX1-2
|
NK1 homeobox 2
|