Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
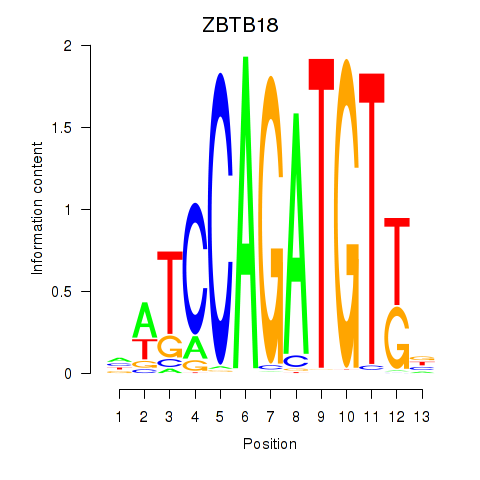
Results for ZBTB18
Z-value: 0.61
Transcription factors associated with ZBTB18
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB18
|
ENSG00000179456.10 | ZBTB18 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB18 | hg38_v1_chr1_+_244051275_244051291 | -0.27 | 1.9e-01 | Click! |
Activity profile of ZBTB18 motif
Sorted Z-values of ZBTB18 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB18
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_119354771 | 1.25 |
ENST00000503646.1
|
TNFAIP8
|
TNF alpha induced protein 8 |
| chr17_+_34270213 | 1.04 |
ENST00000378569.2
ENST00000394627.5 ENST00000394630.3 |
CCL7
|
C-C motif chemokine ligand 7 |
| chr2_+_151357583 | 0.88 |
ENST00000243347.5
|
TNFAIP6
|
TNF alpha induced protein 6 |
| chr2_+_201129318 | 0.76 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD like apoptosis regulator |
| chrX_+_150983350 | 0.68 |
ENST00000455596.5
ENST00000448905.6 |
HMGB3
|
high mobility group box 3 |
| chr15_-_56918571 | 0.67 |
ENST00000559000.6
|
ENSG00000285253.1
|
novel protein |
| chr1_-_112618204 | 0.50 |
ENST00000369664.1
|
ST7L
|
suppression of tumorigenicity 7 like |
| chrX_+_9912434 | 0.49 |
ENST00000418909.6
|
SHROOM2
|
shroom family member 2 |
| chr13_+_97222296 | 0.48 |
ENST00000343600.8
ENST00000376673.7 ENST00000679496.1 ENST00000345429.10 |
MBNL2
|
muscleblind like splicing regulator 2 |
| chr12_+_25973748 | 0.44 |
ENST00000542865.5
|
RASSF8
|
Ras association domain family member 8 |
| chr12_-_104050112 | 0.39 |
ENST00000547583.1
ENST00000546851.1 ENST00000360814.9 |
GLT8D2
|
glycosyltransferase 8 domain containing 2 |
| chr16_-_49664225 | 0.39 |
ENST00000535559.5
|
ZNF423
|
zinc finger protein 423 |
| chr17_+_34255274 | 0.38 |
ENST00000580907.5
ENST00000225831.4 |
CCL2
|
C-C motif chemokine ligand 2 |
| chr2_-_88992903 | 0.38 |
ENST00000495489.1
|
IGKV1-8
|
immunoglobulin kappa variable 1-8 |
| chr19_+_39412650 | 0.37 |
ENST00000425673.6
|
PLEKHG2
|
pleckstrin homology and RhoGEF domain containing G2 |
| chr7_-_36985060 | 0.36 |
ENST00000396040.6
|
ELMO1
|
engulfment and cell motility 1 |
| chr22_-_19881163 | 0.35 |
ENST00000485358.5
|
TXNRD2
|
thioredoxin reductase 2 |
| chr19_+_40613416 | 0.35 |
ENST00000599724.5
ENST00000597071.5 |
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chrX_-_112679919 | 0.34 |
ENST00000371968.8
|
LHFPL1
|
LHFPL tetraspan subfamily member 1 |
| chr17_-_69141878 | 0.33 |
ENST00000590645.1
ENST00000284425.7 |
ABCA6
|
ATP binding cassette subfamily A member 6 |
| chr16_+_24256313 | 0.33 |
ENST00000005284.4
|
CACNG3
|
calcium voltage-gated channel auxiliary subunit gamma 3 |
| chr22_-_19881369 | 0.33 |
ENST00000462330.5
|
TXNRD2
|
thioredoxin reductase 2 |
| chr9_-_13165442 | 0.32 |
ENST00000542239.1
ENST00000538841.5 ENST00000433359.6 |
MPDZ
|
multiple PDZ domain crumbs cell polarity complex component |
| chr14_-_23057530 | 0.29 |
ENST00000397359.7
ENST00000610348.1 |
CDH24
|
cadherin 24 |
| chrX_+_65588368 | 0.29 |
ENST00000609672.5
|
MSN
|
moesin |
| chr3_+_30606574 | 0.28 |
ENST00000295754.10
ENST00000359013.4 |
TGFBR2
|
transforming growth factor beta receptor 2 |
| chr2_-_88966767 | 0.28 |
ENST00000464162.1
|
IGKV1-6
|
immunoglobulin kappa variable 1-6 |
| chr14_-_23057506 | 0.28 |
ENST00000487137.7
|
CDH24
|
cadherin 24 |
| chr5_-_151924846 | 0.27 |
ENST00000274576.9
|
GLRA1
|
glycine receptor alpha 1 |
| chr7_+_101127095 | 0.27 |
ENST00000223095.5
|
SERPINE1
|
serpin family E member 1 |
| chr2_+_188974364 | 0.27 |
ENST00000304636.9
ENST00000317840.9 |
COL3A1
|
collagen type III alpha 1 chain |
| chr18_+_3448456 | 0.27 |
ENST00000549780.5
|
TGIF1
|
TGFB induced factor homeobox 1 |
| chr19_+_11538844 | 0.26 |
ENST00000252456.7
|
CNN1
|
calponin 1 |
| chr8_-_17697654 | 0.26 |
ENST00000297488.10
|
MTUS1
|
microtubule associated scaffold protein 1 |
| chr17_+_34285734 | 0.26 |
ENST00000305869.4
|
CCL11
|
C-C motif chemokine ligand 11 |
| chr5_-_151924824 | 0.26 |
ENST00000455880.2
|
GLRA1
|
glycine receptor alpha 1 |
| chrX_+_150983299 | 0.25 |
ENST00000325307.12
|
HMGB3
|
high mobility group box 3 |
| chr9_+_113501359 | 0.25 |
ENST00000343817.9
ENST00000394646.7 |
RGS3
|
regulator of G protein signaling 3 |
| chr15_+_31326807 | 0.25 |
ENST00000307145.4
|
KLF13
|
Kruppel like factor 13 |
| chr3_-_37174578 | 0.25 |
ENST00000336686.9
|
LRRFIP2
|
LRR binding FLII interacting protein 2 |
| chr17_-_33293247 | 0.24 |
ENST00000225823.7
|
ASIC2
|
acid sensing ion channel subunit 2 |
| chr2_+_33134620 | 0.24 |
ENST00000402934.5
ENST00000404525.5 ENST00000407925.5 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr16_-_29862890 | 0.23 |
ENST00000563415.1
|
CDIPT
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase |
| chrX_+_103776493 | 0.22 |
ENST00000433491.5
ENST00000612423.4 ENST00000443502.5 |
PLP1
|
proteolipid protein 1 |
| chrX_+_103776831 | 0.22 |
ENST00000621218.5
ENST00000619236.1 |
PLP1
|
proteolipid protein 1 |
| chr6_-_30556477 | 0.22 |
ENST00000376621.8
|
GNL1
|
G protein nucleolar 1 (putative) |
| chr5_+_132873660 | 0.22 |
ENST00000296877.3
|
LEAP2
|
liver enriched antimicrobial peptide 2 |
| chr3_-_44510602 | 0.22 |
ENST00000436261.6
|
ZNF852
|
zinc finger protein 852 |
| chr2_+_33134579 | 0.21 |
ENST00000418533.6
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr12_-_53727476 | 0.21 |
ENST00000549784.5
ENST00000262059.8 |
CALCOCO1
|
calcium binding and coiled-coil domain 1 |
| chr2_-_229232039 | 0.19 |
ENST00000354069.6
|
PID1
|
phosphotyrosine interaction domain containing 1 |
| chr10_-_93482194 | 0.19 |
ENST00000358334.9
ENST00000371488.3 |
MYOF
|
myoferlin |
| chr2_-_89117844 | 0.19 |
ENST00000490686.1
|
IGKV1-17
|
immunoglobulin kappa variable 1-17 |
| chr1_-_16980607 | 0.19 |
ENST00000375535.4
|
MFAP2
|
microfibril associated protein 2 |
| chr12_-_47725483 | 0.18 |
ENST00000422538.8
|
ENDOU
|
endonuclease, poly(U) specific |
| chr5_-_149944744 | 0.18 |
ENST00000255266.10
ENST00000617647.4 ENST00000613228.1 |
PDE6A
|
phosphodiesterase 6A |
| chr22_+_20917398 | 0.18 |
ENST00000354336.8
|
CRKL
|
CRK like proto-oncogene, adaptor protein |
| chr12_-_47725558 | 0.18 |
ENST00000229003.7
|
ENDOU
|
endonuclease, poly(U) specific |
| chr11_+_65638085 | 0.18 |
ENST00000534313.6
ENST00000533361.1 ENST00000526137.1 |
SIPA1
|
signal-induced proliferation-associated 1 |
| chr14_-_91244669 | 0.17 |
ENST00000650645.1
|
GPR68
|
G protein-coupled receptor 68 |
| chr19_+_11538767 | 0.17 |
ENST00000592923.5
ENST00000535659.6 |
CNN1
|
calponin 1 |
| chr15_-_55319107 | 0.16 |
ENST00000565225.1
ENST00000436697.3 ENST00000567948.1 ENST00000563262.5 |
PIGBOS1
RAB27A
|
PIGB opposite strand 1 RAB27A, member RAS oncogene family |
| chr17_-_15265230 | 0.16 |
ENST00000676161.1
ENST00000646419.2 ENST00000312280.9 ENST00000494511.7 ENST00000580584.3 ENST00000676221.1 |
PMP22
|
peripheral myelin protein 22 |
| chr11_-_119381629 | 0.16 |
ENST00000260187.7
ENST00000455332.6 |
USP2
|
ubiquitin specific peptidase 2 |
| chr15_-_31160954 | 0.16 |
ENST00000558445.5
ENST00000559177.5 |
TRPM1
|
transient receptor potential cation channel subfamily M member 1 |
| chr10_+_103493931 | 0.16 |
ENST00000369780.8
|
NEURL1
|
neuralized E3 ubiquitin protein ligase 1 |
| chr9_+_94726657 | 0.16 |
ENST00000375315.8
ENST00000277198.6 ENST00000297979.9 |
AOPEP
|
aminopeptidase O (putative) |
| chr11_-_62556230 | 0.15 |
ENST00000530285.5
|
AHNAK
|
AHNAK nucleoprotein |
| chr4_-_104494882 | 0.15 |
ENST00000394767.3
|
CXXC4
|
CXXC finger protein 4 |
| chrX_+_150361559 | 0.15 |
ENST00000262858.8
|
MAMLD1
|
mastermind like domain containing 1 |
| chr1_+_53062052 | 0.15 |
ENST00000395871.7
ENST00000673702.1 ENST00000673956.1 ENST00000312553.10 ENST00000371500.8 ENST00000618387.1 |
PODN
|
podocan |
| chr19_+_50432885 | 0.15 |
ENST00000357701.6
|
MYBPC2
|
myosin binding protein C2 |
| chr6_+_112236806 | 0.15 |
ENST00000588837.5
ENST00000590293.5 ENST00000585450.5 ENST00000629766.2 ENST00000590804.5 ENST00000590584.4 ENST00000628122.2 ENST00000627025.1 ENST00000590673.5 ENST00000585611.5 ENST00000587816.2 |
LAMA4-AS1
ENSG00000281613.2
|
LAMA4 antisense RNA 1 novel ret finger protein-like 4B |
| chr11_+_5389377 | 0.15 |
ENST00000328611.5
|
OR51M1
|
olfactory receptor family 51 subfamily M member 1 |
| chr14_-_38256074 | 0.15 |
ENST00000342213.3
|
CLEC14A
|
C-type lectin domain containing 14A |
| chr12_+_52079700 | 0.14 |
ENST00000546390.2
|
SMIM41
|
small integral membrane protein 41 |
| chr11_-_40294089 | 0.14 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr19_-_10231293 | 0.14 |
ENST00000646641.1
|
S1PR2
|
sphingosine-1-phosphate receptor 2 |
| chr10_+_70052582 | 0.14 |
ENST00000676699.1
|
MACROH2A2
|
macroH2A.2 histone |
| chr6_-_165663170 | 0.14 |
ENST00000539869.4
|
PDE10A
|
phosphodiesterase 10A |
| chr1_-_153545793 | 0.13 |
ENST00000354332.8
ENST00000368716.9 |
S100A4
|
S100 calcium binding protein A4 |
| chr12_+_43836043 | 0.13 |
ENST00000266534.8
ENST00000551577.5 |
TMEM117
|
transmembrane protein 117 |
| chr3_+_49007062 | 0.13 |
ENST00000395474.7
ENST00000610967.4 ENST00000429900.6 |
WDR6
|
WD repeat domain 6 |
| chr16_+_6019585 | 0.13 |
ENST00000547372.5
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr4_+_15374541 | 0.13 |
ENST00000382383.7
ENST00000429690.5 |
C1QTNF7
|
C1q and TNF related 7 |
| chrX_-_102516714 | 0.13 |
ENST00000289373.5
|
TMSB15A
|
thymosin beta 15A |
| chr19_+_9140377 | 0.13 |
ENST00000360385.7
ENST00000247956.11 |
ZNF317
|
zinc finger protein 317 |
| chr7_-_120857124 | 0.12 |
ENST00000441017.5
ENST00000424710.5 ENST00000433758.5 |
TSPAN12
|
tetraspanin 12 |
| chr20_+_59300703 | 0.12 |
ENST00000395654.3
|
EDN3
|
endothelin 3 |
| chr19_-_42302292 | 0.12 |
ENST00000594989.5
|
PAFAH1B3
|
platelet activating factor acetylhydrolase 1b catalytic subunit 3 |
| chr20_+_59300589 | 0.12 |
ENST00000337938.7
ENST00000371025.7 |
EDN3
|
endothelin 3 |
| chr3_+_121593363 | 0.11 |
ENST00000338040.6
|
FBXO40
|
F-box protein 40 |
| chr1_+_42153399 | 0.11 |
ENST00000372581.2
|
GUCA2B
|
guanylate cyclase activator 2B |
| chr9_+_97412062 | 0.11 |
ENST00000355295.5
|
TDRD7
|
tudor domain containing 7 |
| chr14_+_64504743 | 0.11 |
ENST00000683701.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr12_+_112125531 | 0.11 |
ENST00000549358.5
ENST00000257604.9 ENST00000548092.5 ENST00000412615.7 ENST00000552896.1 |
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr15_+_51377247 | 0.11 |
ENST00000396399.6
|
GLDN
|
gliomedin |
| chr17_+_49575828 | 0.10 |
ENST00000328741.6
|
NXPH3
|
neurexophilin 3 |
| chrX_-_111270474 | 0.10 |
ENST00000324068.2
|
CAPN6
|
calpain 6 |
| chr11_-_57322197 | 0.10 |
ENST00000532437.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1 |
| chr19_-_15808126 | 0.10 |
ENST00000334920.3
|
OR10H1
|
olfactory receptor family 10 subfamily H member 1 |
| chr17_-_19748341 | 0.10 |
ENST00000395555.7
|
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1 |
| chr17_-_19748285 | 0.09 |
ENST00000570414.1
ENST00000225740.11 |
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1 |
| chr3_+_124584625 | 0.09 |
ENST00000291478.9
ENST00000682363.1 ENST00000454902.1 |
KALRN
|
kalirin RhoGEF kinase |
| chr20_+_59300547 | 0.09 |
ENST00000644821.1
|
EDN3
|
endothelin 3 |
| chr20_+_59300402 | 0.09 |
ENST00000311585.11
ENST00000371028.6 |
EDN3
|
endothelin 3 |
| chr17_-_19748355 | 0.09 |
ENST00000494157.6
|
ALDH3A1
|
aldehyde dehydrogenase 3 family member A1 |
| chr18_+_54828406 | 0.09 |
ENST00000262094.10
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr12_-_11022620 | 0.09 |
ENST00000390673.2
|
TAS2R19
|
taste 2 receptor member 19 |
| chr11_-_69013233 | 0.09 |
ENST00000309099.7
ENST00000320913.6 |
MRGPRF
|
MAS related GPR family member F |
| chr11_+_73289403 | 0.09 |
ENST00000535931.2
ENST00000544437.6 |
P2RY6
|
pyrimidinergic receptor P2Y6 |
| chr19_-_35510384 | 0.09 |
ENST00000602679.5
ENST00000492341.6 ENST00000472252.6 ENST00000602781.5 ENST00000402589.6 ENST00000458071.5 ENST00000436012.5 ENST00000443640.5 ENST00000450261.1 ENST00000467637.5 ENST00000480502.5 ENST00000474928.5 ENST00000414866.6 ENST00000392206.6 ENST00000488892.5 |
DMKN
|
dermokine |
| chr19_-_51645545 | 0.09 |
ENST00000534261.3
|
SIGLEC5
|
sialic acid binding Ig like lectin 5 |
| chr19_+_42302098 | 0.09 |
ENST00000598490.1
ENST00000341747.8 |
PRR19
|
proline rich 19 |
| chr14_+_67240713 | 0.09 |
ENST00000677382.1
|
MPP5
|
membrane palmitoylated protein 5 |
| chr17_+_50560703 | 0.09 |
ENST00000359106.10
|
CACNA1G
|
calcium voltage-gated channel subunit alpha1 G |
| chr4_+_74445302 | 0.08 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chr3_-_160449530 | 0.08 |
ENST00000494486.1
|
TRIM59
|
tripartite motif containing 59 |
| chr14_+_64503943 | 0.08 |
ENST00000556965.1
ENST00000554015.5 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr4_+_155903688 | 0.08 |
ENST00000536354.3
|
TDO2
|
tryptophan 2,3-dioxygenase |
| chr15_-_74208969 | 0.07 |
ENST00000423167.6
ENST00000432245.6 |
STRA6
|
signaling receptor and transporter of retinol STRA6 |
| chr11_+_60056653 | 0.07 |
ENST00000278865.8
|
MS4A3
|
membrane spanning 4-domains A3 |
| chrX_+_48383516 | 0.07 |
ENST00000620320.4
ENST00000595689.3 |
SSX4
|
SSX family member 4 |
| chr12_+_53985783 | 0.07 |
ENST00000513209.1
|
ENSG00000273049.1
|
novel protein, readthrough between HOXC10 and HOXC5 |
| chr20_+_58907981 | 0.07 |
ENST00000656419.1
|
GNAS
|
GNAS complex locus |
| chr17_+_38428456 | 0.07 |
ENST00000622683.5
ENST00000620417.4 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr14_+_67241278 | 0.07 |
ENST00000676464.1
|
MPP5
|
membrane palmitoylated protein 5 |
| chr10_-_69573416 | 0.06 |
ENST00000242462.5
|
NEUROG3
|
neurogenin 3 |
| chr12_-_55727828 | 0.06 |
ENST00000546939.5
|
CD63
|
CD63 molecule |
| chr18_+_24155938 | 0.06 |
ENST00000582229.1
|
CABYR
|
calcium binding tyrosine phosphorylation regulated |
| chr17_+_50561010 | 0.06 |
ENST00000360761.8
ENST00000354983.8 ENST00000352832.9 |
CACNA1G
|
calcium voltage-gated channel subunit alpha1 G |
| chr11_-_69013356 | 0.06 |
ENST00000441623.1
|
MRGPRF
|
MAS related GPR family member F |
| chr5_+_132673983 | 0.06 |
ENST00000622422.1
ENST00000231449.7 ENST00000350025.2 |
IL4
|
interleukin 4 |
| chr18_+_6834473 | 0.05 |
ENST00000581099.5
ENST00000419673.6 ENST00000531294.5 |
ARHGAP28
|
Rho GTPase activating protein 28 |
| chr17_+_44957907 | 0.05 |
ENST00000678938.1
|
NMT1
|
N-myristoyltransferase 1 |
| chr17_+_62627628 | 0.05 |
ENST00000303375.10
|
MRC2
|
mannose receptor C type 2 |
| chrX_+_100644183 | 0.05 |
ENST00000640889.1
ENST00000373004.5 |
SRPX2
|
sushi repeat containing protein X-linked 2 |
| chr1_+_15684284 | 0.04 |
ENST00000375799.8
ENST00000375793.2 |
PLEKHM2
|
pleckstrin homology and RUN domain containing M2 |
| chr17_-_63972918 | 0.04 |
ENST00000435607.3
|
SCN4A
|
sodium voltage-gated channel alpha subunit 4 |
| chr2_-_224569782 | 0.03 |
ENST00000409096.5
|
CUL3
|
cullin 3 |
| chr12_-_55727796 | 0.03 |
ENST00000550776.5
|
CD63
|
CD63 molecule |
| chr2_-_1744442 | 0.03 |
ENST00000433670.5
ENST00000425171.1 ENST00000252804.9 |
PXDN
|
peroxidasin |
| chr4_+_164877164 | 0.03 |
ENST00000507152.6
ENST00000515275.1 |
APELA
|
apelin receptor early endogenous ligand |
| chr9_+_84668485 | 0.03 |
ENST00000359847.4
ENST00000395882.6 ENST00000376208.6 ENST00000376213.6 |
NTRK2
|
neurotrophic receptor tyrosine kinase 2 |
| chr11_+_60056587 | 0.03 |
ENST00000395032.6
ENST00000358152.6 |
MS4A3
|
membrane spanning 4-domains A3 |
| chr4_-_151227881 | 0.03 |
ENST00000652233.1
ENST00000514152.5 |
SH3D19
|
SH3 domain containing 19 |
| chr4_-_86934700 | 0.03 |
ENST00000473559.5
|
ENSG00000285458.1
|
novel protein |
| chr15_+_55319202 | 0.03 |
ENST00000164305.10
ENST00000566999.5 ENST00000539642.5 |
PIGB
|
phosphatidylinositol glycan anchor biosynthesis class B |
| chr9_+_273026 | 0.02 |
ENST00000682249.1
ENST00000453981.5 ENST00000487230.5 ENST00000469391.5 |
DOCK8
|
dedicator of cytokinesis 8 |
| chr2_-_65432591 | 0.02 |
ENST00000356388.9
|
SPRED2
|
sprouty related EVH1 domain containing 2 |
| chr8_-_48921419 | 0.02 |
ENST00000020945.4
|
SNAI2
|
snail family transcriptional repressor 2 |
| chr1_-_16018005 | 0.02 |
ENST00000406363.2
ENST00000411503.5 ENST00000311890.14 ENST00000487046.1 |
HSPB7
|
heat shock protein family B (small) member 7 |
| chr3_+_184176949 | 0.02 |
ENST00000432591.5
ENST00000431779.5 ENST00000621863.4 |
AP2M1
|
adaptor related protein complex 2 subunit mu 1 |
| chr17_+_44349050 | 0.02 |
ENST00000639447.1
|
GRN
|
granulin precursor |
| chr3_+_184176778 | 0.02 |
ENST00000439647.5
|
AP2M1
|
adaptor related protein complex 2 subunit mu 1 |
| chr7_-_135748712 | 0.02 |
ENST00000415751.1
|
FAM180A
|
family with sequence similarity 180 member A |
| chr20_+_58150896 | 0.02 |
ENST00000371168.4
|
C20orf85
|
chromosome 20 open reading frame 85 |
| chr13_-_60013178 | 0.02 |
ENST00000498416.2
ENST00000465066.5 |
DIAPH3
|
diaphanous related formin 3 |
| chr21_-_34511317 | 0.02 |
ENST00000611936.1
ENST00000337385.7 |
KCNE1
|
potassium voltage-gated channel subfamily E regulatory subunit 1 |
| chr11_+_15114912 | 0.02 |
ENST00000379556.8
|
INSC
|
INSC spindle orientation adaptor protein |
| chrX_-_155334580 | 0.01 |
ENST00000369449.7
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr1_+_23743462 | 0.01 |
ENST00000609199.1
|
ELOA
|
elongin A |
| chr11_-_34357994 | 0.01 |
ENST00000435224.3
|
ABTB2
|
ankyrin repeat and BTB domain containing 2 |
| chr1_-_168729187 | 0.01 |
ENST00000367817.4
|
DPT
|
dermatopontin |
| chr16_-_29899245 | 0.01 |
ENST00000537485.5
|
SEZ6L2
|
seizure related 6 homolog like 2 |
| chr11_-_94493798 | 0.01 |
ENST00000538923.1
ENST00000540013.5 ENST00000407439.7 ENST00000323929.8 ENST00000393241.8 |
MRE11
|
MRE11 homolog, double strand break repair nuclease |
| chr3_+_159852933 | 0.01 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr12_+_1629197 | 0.01 |
ENST00000397196.7
|
WNT5B
|
Wnt family member 5B |
| chr10_+_7788162 | 0.00 |
ENST00000356708.12
ENST00000335698.4 |
ATP5F1C
|
ATP synthase F1 subunit gamma |
| chr1_-_156677400 | 0.00 |
ENST00000368223.4
|
NES
|
nestin |
| chr1_+_8945858 | 0.00 |
ENST00000549778.5
ENST00000377443.7 ENST00000480186.7 ENST00000377436.6 ENST00000377442.3 |
CA6
|
carbonic anhydrase 6 |
| chr1_+_26410809 | 0.00 |
ENST00000254231.4
ENST00000326279.11 |
LIN28A
|
lin-28 homolog A |
| chr4_+_47485268 | 0.00 |
ENST00000273859.8
ENST00000504445.1 |
ATP10D
|
ATPase phospholipid transporting 10D (putative) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 0.4 | GO:2000502 | regulation of natural killer cell chemotaxis(GO:2000501) negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.1 | 0.3 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.1 | 0.3 | GO:0001300 | chronological cell aging(GO:0001300) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.5 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.8 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.2 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.1 | 0.4 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.4 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.3 | GO:0035962 | response to interleukin-13(GO:0035962) |
| 0.0 | 0.2 | GO:0090298 | negative regulation of mitochondrial DNA replication(GO:0090298) negative regulation of mitochondrial DNA metabolic process(GO:1901859) negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.3 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.2 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.0 | 0.5 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.2 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.9 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.4 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.2 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.4 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.2 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.2 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.2 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.1 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.1 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.0 | 0.7 | GO:0000305 | response to oxygen radical(GO:0000305) |
| 0.0 | 0.0 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.0 | 0.3 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.4 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.0 | 0.0 | GO:0090134 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.0 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.8 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.6 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.5 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0097229 | sperm end piece(GO:0097229) |
| 0.0 | 0.3 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.3 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.2 | GO:0043218 | compact myelin(GO:0043218) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.5 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.4 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 0.3 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.1 | 0.4 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 0.4 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.3 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.1 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.3 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.7 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.9 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.8 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 1.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.0 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.8 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.5 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |