Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
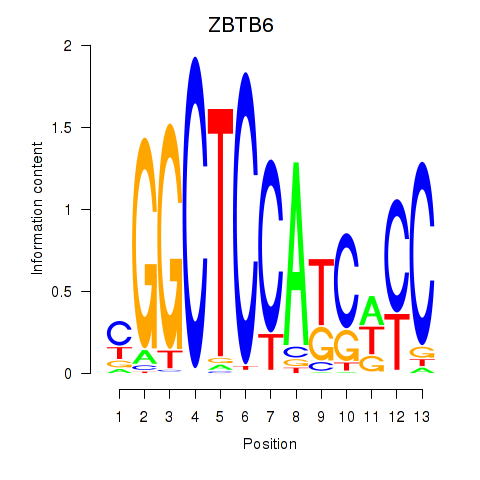
Results for ZBTB6
Z-value: 0.75
Transcription factors associated with ZBTB6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB6
|
ENSG00000186130.5 | ZBTB6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB6 | hg38_v1_chr9_-_122913299_122913333 | -0.01 | 9.5e-01 | Click! |
Activity profile of ZBTB6 motif
Sorted Z-values of ZBTB6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_62248591 | 0.81 |
ENST00000519049.6
|
NKAIN3
|
sodium/potassium transporting ATPase interacting 3 |
| chr3_-_133895753 | 0.79 |
ENST00000460865.3
|
RAB6B
|
RAB6B, member RAS oncogene family |
| chr3_-_116444983 | 0.57 |
ENST00000333617.8
|
LSAMP
|
limbic system associated membrane protein |
| chr1_+_153778178 | 0.54 |
ENST00000532853.5
|
SLC27A3
|
solute carrier family 27 member 3 |
| chr5_-_150302884 | 0.45 |
ENST00000328668.8
|
ARSI
|
arylsulfatase family member I |
| chr12_-_49904217 | 0.44 |
ENST00000550635.6
|
FAIM2
|
Fas apoptotic inhibitory molecule 2 |
| chr22_+_22704265 | 0.42 |
ENST00000390307.2
|
IGLV3-22
|
immunoglobulin lambda variable 3-22 |
| chr6_-_142946312 | 0.40 |
ENST00000367604.6
|
HIVEP2
|
HIVEP zinc finger 2 |
| chr19_+_1507976 | 0.40 |
ENST00000673796.1
|
PLK5
|
polo like kinase 5 (inactive) |
| chr19_-_14979848 | 0.39 |
ENST00000594383.2
|
SLC1A6
|
solute carrier family 1 member 6 |
| chr5_+_170504243 | 0.38 |
ENST00000520740.5
|
KCNIP1
|
potassium voltage-gated channel interacting protein 1 |
| chr5_+_170504005 | 0.37 |
ENST00000328939.9
ENST00000390656.8 |
KCNIP1
|
potassium voltage-gated channel interacting protein 1 |
| chr6_+_137871208 | 0.35 |
ENST00000614035.4
ENST00000621150.3 ENST00000619035.4 ENST00000615468.4 ENST00000620204.3 |
TNFAIP3
|
TNF alpha induced protein 3 |
| chr19_+_12995554 | 0.34 |
ENST00000397661.6
|
NFIX
|
nuclear factor I X |
| chr20_-_25058115 | 0.34 |
ENST00000323482.9
|
ACSS1
|
acyl-CoA synthetase short chain family member 1 |
| chr20_-_25058168 | 0.34 |
ENST00000432802.6
|
ACSS1
|
acyl-CoA synthetase short chain family member 1 |
| chr1_+_26529745 | 0.33 |
ENST00000374168.7
ENST00000374166.8 |
RPS6KA1
|
ribosomal protein S6 kinase A1 |
| chr11_+_111540659 | 0.30 |
ENST00000375615.7
ENST00000525126.5 ENST00000375614.7 ENST00000533265.5 |
LAYN
|
layilin |
| chr1_+_26410809 | 0.30 |
ENST00000254231.4
ENST00000326279.11 |
LIN28A
|
lin-28 homolog A |
| chr11_-_6655788 | 0.30 |
ENST00000299441.5
|
DCHS1
|
dachsous cadherin-related 1 |
| chr16_-_28491759 | 0.29 |
ENST00000565316.6
ENST00000355477.10 ENST00000636228.1 ENST00000333496.14 ENST00000565778.6 ENST00000357806.11 |
CLN3
|
CLN3 lysosomal/endosomal transmembrane protein, battenin |
| chrX_+_49879475 | 0.29 |
ENST00000621775.2
|
USP27X
|
ubiquitin specific peptidase 27 X-linked |
| chr21_+_10521569 | 0.28 |
ENST00000612957.4
ENST00000427445.6 ENST00000612746.1 ENST00000618007.5 |
TPTE
|
transmembrane phosphatase with tensin homology |
| chr22_-_30387334 | 0.28 |
ENST00000382363.8
|
RNF215
|
ring finger protein 215 |
| chr6_+_154995258 | 0.28 |
ENST00000682666.1
|
TIAM2
|
TIAM Rac1 associated GEF 2 |
| chr16_-_11587162 | 0.28 |
ENST00000570904.5
ENST00000574701.5 |
LITAF
|
lipopolysaccharide induced TNF factor |
| chr10_-_103855406 | 0.27 |
ENST00000355946.6
ENST00000369774.8 |
SH3PXD2A
|
SH3 and PX domains 2A |
| chr4_+_663696 | 0.27 |
ENST00000471824.6
|
PDE6B
|
phosphodiesterase 6B |
| chr12_-_6700377 | 0.27 |
ENST00000540656.5
|
PIANP
|
PILR alpha associated neural protein |
| chr14_+_73612517 | 0.27 |
ENST00000645972.2
|
ACOT6
|
acyl-CoA thioesterase 6 |
| chr16_-_30113528 | 0.26 |
ENST00000406256.8
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr16_-_28491996 | 0.26 |
ENST00000568497.6
ENST00000357857.14 ENST00000395653.9 ENST00000635973.1 ENST00000568443.2 ENST00000568558.6 ENST00000565688.5 ENST00000637100.1 ENST00000359984.12 ENST00000636147.2 ENST00000567963.6 |
CLN3
|
CLN3 lysosomal/endosomal transmembrane protein, battenin |
| chrX_+_30235894 | 0.26 |
ENST00000620842.1
|
MAGEB3
|
MAGE family member B3 |
| chr10_-_62268837 | 0.26 |
ENST00000373789.8
|
RTKN2
|
rhotekin 2 |
| chr9_-_114387973 | 0.26 |
ENST00000374088.8
|
AKNA
|
AT-hook transcription factor |
| chr12_-_52493250 | 0.25 |
ENST00000330722.7
|
KRT6A
|
keratin 6A |
| chr8_+_62248933 | 0.25 |
ENST00000523211.5
ENST00000524201.1 |
NKAIN3
|
sodium/potassium transporting ATPase interacting 3 |
| chr22_-_30387078 | 0.25 |
ENST00000215798.10
|
RNF215
|
ring finger protein 215 |
| chr22_-_30574654 | 0.25 |
ENST00000406361.6
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr7_-_105679089 | 0.25 |
ENST00000477775.5
|
ATXN7L1
|
ataxin 7 like 1 |
| chr21_+_10521536 | 0.24 |
ENST00000622113.4
|
TPTE
|
transmembrane phosphatase with tensin homology |
| chr16_-_11586903 | 0.24 |
ENST00000571459.5
ENST00000570798.5 ENST00000622633.5 ENST00000572255.5 ENST00000574763.5 ENST00000574703.5 ENST00000571277.1 |
LITAF
|
lipopolysaccharide induced TNF factor |
| chr3_+_112333147 | 0.24 |
ENST00000473539.5
ENST00000315711.12 ENST00000383681.7 |
CD200
|
CD200 molecule |
| chr18_-_50195138 | 0.24 |
ENST00000285039.12
|
MYO5B
|
myosin VB |
| chr9_-_130939205 | 0.24 |
ENST00000372338.9
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr16_-_11586941 | 0.24 |
ENST00000571976.1
ENST00000413364.6 |
LITAF
|
lipopolysaccharide induced TNF factor |
| chr1_-_45500040 | 0.24 |
ENST00000629482.3
|
CCDC163
|
coiled-coil domain containing 163 |
| chr17_+_35731620 | 0.24 |
ENST00000603017.2
|
RASL10B
|
RAS like family 10 member B |
| chr7_+_118214633 | 0.23 |
ENST00000477532.5
|
ANKRD7
|
ankyrin repeat domain 7 |
| chr18_+_65751000 | 0.23 |
ENST00000397968.4
|
CDH7
|
cadherin 7 |
| chr12_-_6700788 | 0.23 |
ENST00000320591.9
ENST00000534837.6 |
PIANP
|
PILR alpha associated neural protein |
| chr7_-_108240049 | 0.23 |
ENST00000379022.8
|
NRCAM
|
neuronal cell adhesion molecule |
| chr9_-_113299765 | 0.22 |
ENST00000478815.1
|
RNF183
|
ring finger protein 183 |
| chr7_+_144355288 | 0.22 |
ENST00000498580.5
ENST00000056217.10 |
ARHGEF5
|
Rho guanine nucleotide exchange factor 5 |
| chr12_-_6607397 | 0.22 |
ENST00000645005.1
ENST00000646806.1 |
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr19_+_53554819 | 0.22 |
ENST00000505949.5
ENST00000513265.5 ENST00000648511.1 |
ZNF331
|
zinc finger protein 331 |
| chr2_-_27380715 | 0.22 |
ENST00000323703.11
ENST00000436006.1 |
ZNF513
|
zinc finger protein 513 |
| chr20_+_62817023 | 0.22 |
ENST00000649368.1
|
COL9A3
|
collagen type IX alpha 3 chain |
| chr10_+_115093404 | 0.22 |
ENST00000527407.5
|
ATRNL1
|
attractin like 1 |
| chr2_+_85133376 | 0.22 |
ENST00000282111.4
|
TCF7L1
|
transcription factor 7 like 1 |
| chr22_-_30574572 | 0.21 |
ENST00000402369.5
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr17_-_81637049 | 0.21 |
ENST00000374747.9
ENST00000331134.11 |
NPLOC4
|
NPL4 homolog, ubiquitin recognition factor |
| chr16_+_50742110 | 0.21 |
ENST00000566679.6
ENST00000564634.5 ENST00000398568.6 |
CYLD
|
CYLD lysine 63 deubiquitinase |
| chr1_+_110034607 | 0.21 |
ENST00000369795.8
|
STRIP1
|
striatin interacting protein 1 |
| chr1_+_220879434 | 0.21 |
ENST00000366903.8
|
HLX
|
H2.0 like homeobox |
| chr5_-_139904460 | 0.21 |
ENST00000340391.8
|
NRG2
|
neuregulin 2 |
| chr16_+_50742037 | 0.21 |
ENST00000569418.5
|
CYLD
|
CYLD lysine 63 deubiquitinase |
| chr19_+_43716095 | 0.21 |
ENST00000596627.1
|
IRGC
|
immunity related GTPase cinema |
| chr1_-_154961720 | 0.21 |
ENST00000368457.3
|
PYGO2
|
pygopus family PHD finger 2 |
| chr1_-_204196482 | 0.20 |
ENST00000367194.5
|
KISS1
|
KiSS-1 metastasis suppressor |
| chr20_-_63831214 | 0.20 |
ENST00000302995.2
ENST00000245663.9 |
ZBTB46
|
zinc finger and BTB domain containing 46 |
| chr19_-_3700390 | 0.20 |
ENST00000679885.1
ENST00000537021.1 ENST00000589578.5 ENST00000539785.5 ENST00000335312.8 |
PIP5K1C
|
phosphatidylinositol-4-phosphate 5-kinase type 1 gamma |
| chr12_-_6607334 | 0.20 |
ENST00000645645.1
ENST00000357008.7 ENST00000544484.6 ENST00000544040.7 |
CHD4
|
chromodomain helicase DNA binding protein 4 |
| chr19_+_4304588 | 0.20 |
ENST00000221856.11
|
FSD1
|
fibronectin type III and SPRY domain containing 1 |
| chr9_-_124771238 | 0.20 |
ENST00000344523.8
ENST00000373584.7 |
NR6A1
|
nuclear receptor subfamily 6 group A member 1 |
| chr17_-_1649854 | 0.20 |
ENST00000301336.7
|
RILP
|
Rab interacting lysosomal protein |
| chr11_+_5351508 | 0.20 |
ENST00000380219.1
|
OR51B6
|
olfactory receptor family 51 subfamily B member 6 |
| chr11_-_66546021 | 0.19 |
ENST00000310442.5
|
ZDHHC24
|
zinc finger DHHC-type containing 24 |
| chr2_+_119544420 | 0.19 |
ENST00000413369.8
|
CFAP221
|
cilia and flagella associated protein 221 |
| chr1_+_45500287 | 0.19 |
ENST00000401061.9
ENST00000616135.1 |
MMACHC
|
metabolism of cobalamin associated C |
| chr22_+_22887780 | 0.19 |
ENST00000532223.2
ENST00000526893.6 ENST00000531372.1 |
IGLL5
|
immunoglobulin lambda like polypeptide 5 |
| chr5_+_175871670 | 0.19 |
ENST00000514150.5
|
CPLX2
|
complexin 2 |
| chr8_-_143609547 | 0.18 |
ENST00000433751.5
ENST00000495276.6 ENST00000220966.10 |
PYCR3
|
pyrroline-5-carboxylate reductase 3 |
| chr1_-_44843240 | 0.18 |
ENST00000372192.4
|
PTCH2
|
patched 2 |
| chr19_-_38426162 | 0.18 |
ENST00000587738.2
ENST00000586305.5 |
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr19_+_51127030 | 0.18 |
ENST00000599948.1
|
SIGLEC9
|
sialic acid binding Ig like lectin 9 |
| chr3_-_8769602 | 0.18 |
ENST00000316793.8
ENST00000431493.1 |
OXTR
|
oxytocin receptor |
| chr1_+_14945775 | 0.18 |
ENST00000400797.3
|
KAZN
|
kazrin, periplakin interacting protein |
| chr11_-_111766359 | 0.17 |
ENST00000341980.10
ENST00000311129.9 ENST00000393055.6 ENST00000426998.6 ENST00000527614.6 |
PPP2R1B
|
protein phosphatase 2 scaffold subunit Abeta |
| chr4_+_653171 | 0.17 |
ENST00000488061.5
ENST00000429163.6 |
PDE6B
|
phosphodiesterase 6B |
| chr12_+_6821646 | 0.17 |
ENST00000428545.6
|
GPR162
|
G protein-coupled receptor 162 |
| chr17_+_49495286 | 0.17 |
ENST00000172229.8
|
NGFR
|
nerve growth factor receptor |
| chr11_-_40294089 | 0.17 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr20_-_23879735 | 0.17 |
ENST00000304710.5
|
CST5
|
cystatin D |
| chrY_+_9466955 | 0.17 |
ENST00000423647.6
ENST00000451548.6 |
TSPY1
|
testis specific protein Y-linked 1 |
| chr19_+_12995467 | 0.16 |
ENST00000592199.6
|
NFIX
|
nuclear factor I X |
| chr10_-_119536533 | 0.16 |
ENST00000392865.5
|
RGS10
|
regulator of G protein signaling 10 |
| chr20_+_45857607 | 0.16 |
ENST00000255152.3
|
ZSWIM3
|
zinc finger SWIM-type containing 3 |
| chr3_+_196216527 | 0.16 |
ENST00000296327.10
|
SLC51A
|
solute carrier family 51 subunit alpha |
| chr10_+_70815889 | 0.16 |
ENST00000373202.8
|
SGPL1
|
sphingosine-1-phosphate lyase 1 |
| chrX_-_133985449 | 0.16 |
ENST00000631057.2
|
GPC3
|
glypican 3 |
| chr17_+_40443441 | 0.16 |
ENST00000269593.5
|
IGFBP4
|
insulin like growth factor binding protein 4 |
| chrY_+_9527880 | 0.16 |
ENST00000428845.6
ENST00000444056.1 |
TSPY10
|
testis specific protein Y-linked 10 |
| chr19_+_43716070 | 0.16 |
ENST00000244314.6
|
IRGC
|
immunity related GTPase cinema |
| chr8_+_55102448 | 0.16 |
ENST00000622811.1
|
XKR4
|
XK related 4 |
| chr6_-_36839434 | 0.16 |
ENST00000244751.7
ENST00000633280.1 |
CPNE5
|
copine 5 |
| chr17_+_7252268 | 0.16 |
ENST00000396628.6
ENST00000574993.5 ENST00000573657.5 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr17_-_28357455 | 0.16 |
ENST00000618887.2
ENST00000540200.6 |
POLDIP2
|
DNA polymerase delta interacting protein 2 |
| chr19_-_51417700 | 0.15 |
ENST00000529627.1
ENST00000439889.6 |
SIGLEC10
|
sialic acid binding Ig like lectin 10 |
| chrY_+_9357797 | 0.15 |
ENST00000287721.13
ENST00000383005.7 ENST00000383000.1 |
TSPY8
|
testis specific protein Y-linked 8 |
| chr12_+_6821797 | 0.15 |
ENST00000311268.8
ENST00000382315.7 |
GPR162
|
G protein-coupled receptor 162 |
| chr17_-_7329266 | 0.15 |
ENST00000571887.5
ENST00000315614.11 ENST00000399464.7 ENST00000570460.5 |
NEURL4
|
neuralized E3 ubiquitin protein ligase 4 |
| chrY_+_6246223 | 0.15 |
ENST00000429039.5
ENST00000320701.8 ENST00000383042.1 |
TSPY2
|
testis specific protein Y-linked 2 |
| chr14_-_60724300 | 0.15 |
ENST00000556952.3
ENST00000216513.5 |
SIX4
|
SIX homeobox 4 |
| chr17_+_7252237 | 0.15 |
ENST00000570500.5
|
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr11_-_6234618 | 0.15 |
ENST00000449352.7
|
FAM160A2
|
family with sequence similarity 160 member A2 |
| chr22_-_41915053 | 0.15 |
ENST00000621082.2
|
SHISA8
|
shisa family member 8 |
| chr19_+_7507082 | 0.15 |
ENST00000599312.1
|
ENSG00000267952.1
|
novel transcript |
| chr19_-_38426195 | 0.15 |
ENST00000615439.5
ENST00000614135.4 ENST00000622174.4 ENST00000587753.5 ENST00000454404.6 ENST00000617966.4 ENST00000618320.4 ENST00000293062.13 ENST00000433821.6 ENST00000426920.6 |
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr4_-_119628791 | 0.15 |
ENST00000354960.8
|
PDE5A
|
phosphodiesterase 5A |
| chr2_+_218859794 | 0.15 |
ENST00000233948.4
|
WNT6
|
Wnt family member 6 |
| chr11_+_47980538 | 0.15 |
ENST00000613246.4
ENST00000418331.7 ENST00000615445.4 ENST00000440289.6 |
PTPRJ
|
protein tyrosine phosphatase receptor type J |
| chr14_-_23302823 | 0.15 |
ENST00000452015.9
|
PPP1R3E
|
protein phosphatase 1 regulatory subunit 3E |
| chr10_+_47322450 | 0.14 |
ENST00000581492.3
|
GDF2
|
growth differentiation factor 2 |
| chr2_-_152099023 | 0.14 |
ENST00000201943.10
ENST00000427385.6 ENST00000539935.7 |
CACNB4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr17_-_7252054 | 0.14 |
ENST00000575783.5
ENST00000573600.5 |
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr11_-_6234653 | 0.14 |
ENST00000524416.1
ENST00000265978.8 |
FAM160A2
|
family with sequence similarity 160 member A2 |
| chr9_-_133129395 | 0.14 |
ENST00000393157.8
|
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr11_+_45922640 | 0.14 |
ENST00000401752.6
ENST00000325468.9 |
LARGE2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr4_-_138242325 | 0.14 |
ENST00000280612.9
|
SLC7A11
|
solute carrier family 7 member 11 |
| chr7_-_151167692 | 0.14 |
ENST00000297537.5
|
GBX1
|
gastrulation brain homeobox 1 |
| chrY_+_9337464 | 0.14 |
ENST00000426950.6
ENST00000640033.1 ENST00000383008.1 |
TSPY4
|
testis specific protein Y-linked 4 |
| chr3_+_19148500 | 0.14 |
ENST00000328405.7
|
KCNH8
|
potassium voltage-gated channel subfamily H member 8 |
| chr1_-_54887161 | 0.14 |
ENST00000535035.6
ENST00000371269.9 ENST00000436604.2 |
DHCR24
|
24-dehydrocholesterol reductase |
| chr3_+_160225409 | 0.14 |
ENST00000326474.5
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr6_-_114343012 | 0.14 |
ENST00000312719.10
|
HS3ST5
|
heparan sulfate-glucosamine 3-sulfotransferase 5 |
| chr15_+_70892809 | 0.14 |
ENST00000260382.10
ENST00000560755.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr1_+_40161355 | 0.14 |
ENST00000372771.5
|
RLF
|
RLF zinc finger |
| chrX_-_75785205 | 0.14 |
ENST00000373359.4
|
MAGEE2
|
MAGE family member E2 |
| chr1_+_1033987 | 0.14 |
ENST00000651234.1
ENST00000652369.1 |
AGRN
|
agrin |
| chr13_+_98143410 | 0.14 |
ENST00000596580.2
ENST00000376581.9 |
FARP1
|
FERM, ARH/RhoGEF and pleckstrin domain protein 1 |
| chr14_+_100128573 | 0.14 |
ENST00000554695.5
|
EVL
|
Enah/Vasp-like |
| chr5_-_148654522 | 0.14 |
ENST00000377888.8
|
HTR4
|
5-hydroxytryptamine receptor 4 |
| chr1_-_156751597 | 0.14 |
ENST00000537739.5
|
HDGF
|
heparin binding growth factor |
| chr8_+_123416735 | 0.14 |
ENST00000524254.5
|
NTAQ1
|
N-terminal glutamine amidase 1 |
| chr19_+_53538415 | 0.14 |
ENST00000648122.1
|
ZNF331
|
zinc finger protein 331 |
| chr9_+_127716073 | 0.14 |
ENST00000373289.4
|
TTC16
|
tetratricopeptide repeat domain 16 |
| chrX_-_31072035 | 0.14 |
ENST00000359202.5
|
FTHL17
|
ferritin heavy chain like 17 |
| chr9_-_120929160 | 0.14 |
ENST00000540010.1
|
TRAF1
|
TNF receptor associated factor 1 |
| chr17_-_29930062 | 0.14 |
ENST00000579954.1
ENST00000269033.7 ENST00000540801.6 ENST00000590153.1 ENST00000582084.1 |
SSH2
|
slingshot protein phosphatase 2 |
| chr17_+_42536510 | 0.14 |
ENST00000585572.1
ENST00000586516.5 ENST00000591587.1 |
ENSG00000266929.1
NAGLU
|
novel transcript N-acetyl-alpha-glucosaminidase |
| chr16_-_81077078 | 0.14 |
ENST00000565253.1
ENST00000378611.8 ENST00000299578.10 |
C16orf46
|
chromosome 16 open reading frame 46 |
| chr10_+_115093331 | 0.14 |
ENST00000609571.5
ENST00000355044.8 ENST00000526946.5 |
ATRNL1
|
attractin like 1 |
| chr19_+_50649445 | 0.13 |
ENST00000425202.6
|
C19orf81
|
chromosome 19 open reading frame 81 |
| chr14_-_102509713 | 0.13 |
ENST00000286918.9
|
ANKRD9
|
ankyrin repeat domain 9 |
| chr16_-_30893942 | 0.13 |
ENST00000572628.5
|
BCL7C
|
BAF chromatin remodeling complex subunit BCL7C |
| chr10_-_689613 | 0.13 |
ENST00000280886.12
ENST00000634311.1 |
DIP2C
|
disco interacting protein 2 homolog C |
| chr17_-_45262084 | 0.13 |
ENST00000331780.5
|
SPATA32
|
spermatogenesis associated 32 |
| chr16_-_30893990 | 0.13 |
ENST00000215115.5
|
BCL7C
|
BAF chromatin remodeling complex subunit BCL7C |
| chr19_+_55075862 | 0.13 |
ENST00000201647.11
|
EPS8L1
|
EPS8 like 1 |
| chr19_-_55166671 | 0.13 |
ENST00000455045.5
|
DNAAF3
|
dynein axonemal assembly factor 3 |
| chr22_-_38844020 | 0.13 |
ENST00000333039.4
|
NPTXR
|
neuronal pentraxin receptor |
| chr17_+_4584519 | 0.13 |
ENST00000389313.9
|
SMTNL2
|
smoothelin like 2 |
| chr9_-_114657791 | 0.13 |
ENST00000423632.3
|
TEX53
|
testis expressed 53 |
| chr10_+_86958557 | 0.13 |
ENST00000372017.4
ENST00000348795.8 |
SNCG
|
synuclein gamma |
| chr6_+_150143018 | 0.13 |
ENST00000361131.5
|
PPP1R14C
|
protein phosphatase 1 regulatory inhibitor subunit 14C |
| chr3_-_196968822 | 0.12 |
ENST00000412723.6
|
PIGZ
|
phosphatidylinositol glycan anchor biosynthesis class Z |
| chr19_+_4304632 | 0.12 |
ENST00000597590.5
|
FSD1
|
fibronectin type III and SPRY domain containing 1 |
| chr19_-_4581755 | 0.12 |
ENST00000676793.1
|
SEMA6B
|
semaphorin 6B |
| chr2_+_219279330 | 0.12 |
ENST00000425450.5
ENST00000392086.8 ENST00000421532.5 ENST00000336576.10 |
DNAJB2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr17_-_73644435 | 0.12 |
ENST00000392650.8
|
SDK2
|
sidekick cell adhesion molecule 2 |
| chr2_+_110732539 | 0.12 |
ENST00000676595.1
ENST00000389811.8 ENST00000439055.6 |
ACOXL
|
acyl-CoA oxidase like |
| chr6_-_166308385 | 0.12 |
ENST00000322583.5
|
PRR18
|
proline rich 18 |
| chr5_-_95284535 | 0.12 |
ENST00000515393.5
|
MCTP1
|
multiple C2 and transmembrane domain containing 1 |
| chr1_-_204411804 | 0.12 |
ENST00000367188.5
|
PPP1R15B
|
protein phosphatase 1 regulatory subunit 15B |
| chr22_+_19131271 | 0.12 |
ENST00000399635.4
|
TSSK2
|
testis specific serine kinase 2 |
| chr9_+_122370523 | 0.12 |
ENST00000643810.1
ENST00000540753.6 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 |
| chr6_+_42564060 | 0.12 |
ENST00000372903.6
|
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr12_-_121274510 | 0.12 |
ENST00000392474.6
|
CAMKK2
|
calcium/calmodulin dependent protein kinase kinase 2 |
| chr11_-_7796942 | 0.12 |
ENST00000329434.3
|
OR5P2
|
olfactory receptor family 5 subfamily P member 2 |
| chr9_-_29739 | 0.12 |
ENST00000442898.5
|
WASHC1
|
WASH complex subunit 1 |
| chr19_-_55166632 | 0.12 |
ENST00000532817.5
ENST00000527223.6 ENST00000391720.8 |
DNAAF3
|
dynein axonemal assembly factor 3 |
| chr19_-_19733091 | 0.12 |
ENST00000344099.4
|
ZNF14
|
zinc finger protein 14 |
| chr3_-_190120881 | 0.12 |
ENST00000319332.10
|
P3H2
|
prolyl 3-hydroxylase 2 |
| chr17_+_7252024 | 0.12 |
ENST00000573513.5
ENST00000354429.6 ENST00000574255.5 ENST00000356683.6 ENST00000396627.6 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr19_-_55166565 | 0.12 |
ENST00000526003.5
ENST00000534170.5 ENST00000524407.7 |
DNAAF3
|
dynein axonemal assembly factor 3 |
| chr7_+_139231225 | 0.12 |
ENST00000473989.8
|
UBN2
|
ubinuclein 2 |
| chrX_-_153830527 | 0.12 |
ENST00000393758.7
ENST00000544474.5 |
PDZD4
|
PDZ domain containing 4 |
| chr11_-_62612725 | 0.12 |
ENST00000419857.1
ENST00000394773.7 |
EML3
|
EMAP like 3 |
| chr17_+_42536226 | 0.12 |
ENST00000225927.7
|
NAGLU
|
N-acetyl-alpha-glucosaminidase |
| chr11_+_57542641 | 0.12 |
ENST00000527972.5
ENST00000399154.3 |
SMTNL1
|
smoothelin like 1 |
| chr19_-_49423441 | 0.12 |
ENST00000270631.2
|
PTH2
|
parathyroid hormone 2 |
| chr21_+_45405117 | 0.12 |
ENST00000651438.1
|
COL18A1
|
collagen type XVIII alpha 1 chain |
| chr10_+_80132591 | 0.12 |
ENST00000372267.6
|
PLAC9
|
placenta associated 9 |
| chr12_+_50085194 | 0.12 |
ENST00000381513.8
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr19_-_6424772 | 0.12 |
ENST00000619396.4
ENST00000398148.7 |
KHSRP
|
KH-type splicing regulatory protein |
| chrX_-_153830490 | 0.11 |
ENST00000164640.8
|
PDZD4
|
PDZ domain containing 4 |
| chr7_-_99144053 | 0.11 |
ENST00000361125.1
ENST00000361368.7 |
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1 |
| chr19_+_35268921 | 0.11 |
ENST00000222305.8
ENST00000343550.9 |
USF2
|
upstream transcription factor 2, c-fos interacting |
| chr17_+_36601566 | 0.11 |
ENST00000614766.5
|
MRM1
|
mitochondrial rRNA methyltransferase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0019413 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.2 | 0.6 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 0.5 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 0.3 | GO:0070429 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.1 | 0.3 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.1 | 0.8 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.1 | 0.4 | GO:0002357 | defense response to tumor cell(GO:0002357) |
| 0.1 | 0.2 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.1 | 0.2 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.1 | 0.3 | GO:0051710 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 0.1 | 0.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.2 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.1 | 0.3 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.3 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 0.5 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.2 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.1 | GO:0035048 | splicing factor protein import into nucleus(GO:0035048) |
| 0.0 | 0.1 | GO:0003420 | regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.2 | GO:1903974 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) positive regulation of microglial cell migration(GO:1904141) |
| 0.0 | 0.2 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.3 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.0 | 0.1 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.0 | 0.1 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.0 | 0.3 | GO:0070778 | L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 0.0 | 0.3 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.1 | GO:0090291 | negative regulation of osteoclast proliferation(GO:0090291) |
| 0.0 | 0.1 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.2 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.0 | GO:0072138 | mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 0.0 | 0.2 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.2 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.3 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.4 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.2 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0045629 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.4 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.0 | 0.1 | GO:0019230 | proprioception(GO:0019230) |
| 0.0 | 0.2 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.1 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0072249 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.0 | 0.1 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.1 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.1 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.2 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.2 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:2000909 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.1 | GO:1904692 | positive regulation of type B pancreatic cell proliferation(GO:1904692) |
| 0.0 | 0.1 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.0 | 0.1 | GO:1901536 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.2 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.6 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.1 | GO:1901073 | N-acetylglucosamine biosynthetic process(GO:0006045) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.0 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.7 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.1 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.1 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.2 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.1 | GO:2000830 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0060913 | visceral motor neuron differentiation(GO:0021524) cardiac cell fate determination(GO:0060913) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 0.0 | 0.1 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.2 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.3 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.2 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.2 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.6 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.1 | GO:0090472 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.0 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 0.1 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.1 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.3 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.1 | GO:1903984 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.0 | 0.1 | GO:0061360 | optic nerve formation(GO:0021634) optic chiasma development(GO:0061360) nephric duct formation(GO:0072179) regulation of optic nerve formation(GO:2000595) positive regulation of optic nerve formation(GO:2000597) |
| 0.0 | 0.1 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.1 | GO:1904636 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.0 | 0.0 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.5 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.2 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.0 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.0 | 0.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.1 | GO:0036481 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036481) Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.1 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.0 | GO:0005999 | xylulose biosynthetic process(GO:0005999) |
| 0.0 | 0.1 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 0.0 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.0 | 0.1 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.0 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.2 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.1 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.0 | GO:1901624 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 0.0 | 0.1 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.2 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.1 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0071543 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0098560 | cytoplasmic side of early endosome membrane(GO:0098559) cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 0.2 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.1 | 0.3 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.2 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.2 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.1 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.5 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.0 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.3 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.1 | 1.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0004314 | [acyl-carrier-protein] S-malonyltransferase activity(GO:0004314) 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) S-malonyltransferase activity(GO:0016419) malonyltransferase activity(GO:0016420) |
| 0.0 | 0.5 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.0 | 0.2 | GO:0052812 | phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 0.0 | 0.2 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.1 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.3 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.0 | 0.1 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.1 | GO:0044714 | GTP diphosphatase activity(GO:0036219) 2-hydroxy-adenosine triphosphate pyrophosphatase activity(GO:0044713) 2-hydroxy-(deoxy)adenosine-triphosphate pyrophosphatase activity(GO:0044714) ATP diphosphatase activity(GO:0047693) |
| 0.0 | 0.1 | GO:0052816 | medium-chain acyl-CoA hydrolase activity(GO:0052815) long-chain acyl-CoA hydrolase activity(GO:0052816) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.1 | GO:0045174 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.6 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.0 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.0 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.2 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.0 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.0 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.2 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0052842 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.0 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.3 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.1 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.0 | 0.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |