Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
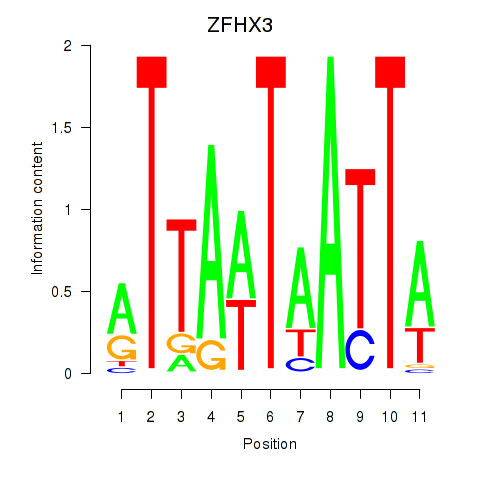
Results for ZFHX3
Z-value: 0.26
Transcription factors associated with ZFHX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZFHX3
|
ENSG00000140836.17 | ZFHX3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZFHX3 | hg38_v1_chr16_-_73048104_73048136 | 0.46 | 2.1e-02 | Click! |
Activity profile of ZFHX3 motif
Sorted Z-values of ZFHX3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZFHX3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_148444674 | 0.38 |
ENST00000344721.8
|
NR3C2
|
nuclear receptor subfamily 3 group C member 2 |
| chrX_-_32155462 | 0.26 |
ENST00000359836.5
ENST00000378707.7 ENST00000541735.5 ENST00000684130.1 ENST00000682238.1 ENST00000620040.5 ENST00000474231.5 |
DMD
|
dystrophin |
| chrM_-_14669 | 0.25 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 6 |
| chr1_-_247758680 | 0.18 |
ENST00000408896.4
|
OR1C1
|
olfactory receptor family 1 subfamily C member 1 |
| chr2_+_87748087 | 0.15 |
ENST00000359481.9
|
PLGLB2
|
plasminogen like B2 |
| chr17_-_41098084 | 0.14 |
ENST00000318329.6
ENST00000333822.5 |
KRTAP4-8
|
keratin associated protein 4-8 |
| chr2_-_87021844 | 0.14 |
ENST00000355705.4
ENST00000409310.6 |
PLGLB1
|
plasminogen like B1 |
| chr7_+_100428782 | 0.14 |
ENST00000414441.5
|
MEPCE
|
methylphosphate capping enzyme |
| chr17_-_69244846 | 0.14 |
ENST00000269081.8
|
ABCA10
|
ATP binding cassette subfamily A member 10 |
| chr6_+_157036315 | 0.13 |
ENST00000637904.1
|
ARID1B
|
AT-rich interaction domain 1B |
| chr2_-_60553558 | 0.13 |
ENST00000642439.1
ENST00000356842.9 |
BCL11A
|
BAF chromatin remodeling complex subunit BCL11A |
| chr22_+_31092447 | 0.13 |
ENST00000455608.5
|
SMTN
|
smoothelin |
| chr6_-_116060859 | 0.13 |
ENST00000606080.2
|
FRK
|
fyn related Src family tyrosine kinase |
| chr22_-_30246739 | 0.12 |
ENST00000403987.3
ENST00000249075.4 |
LIF
|
LIF interleukin 6 family cytokine |
| chr4_-_154590735 | 0.12 |
ENST00000403106.8
ENST00000622532.1 ENST00000651975.1 |
FGA
|
fibrinogen alpha chain |
| chr19_+_926001 | 0.12 |
ENST00000263620.8
|
ARID3A
|
AT-rich interaction domain 3A |
| chr12_-_2835018 | 0.11 |
ENST00000337508.9
|
NRIP2
|
nuclear receptor interacting protein 2 |
| chr14_+_39233884 | 0.11 |
ENST00000553728.1
|
MIA2
|
MIA SH3 domain ER export factor 2 |
| chr4_+_70592253 | 0.10 |
ENST00000322937.10
ENST00000613447.4 |
AMBN
|
ameloblastin |
| chr2_-_2324642 | 0.09 |
ENST00000650485.1
ENST00000649207.1 |
MYT1L
|
myelin transcription factor 1 like |
| chr2_-_2324323 | 0.09 |
ENST00000648339.1
ENST00000647694.1 |
MYT1L
|
myelin transcription factor 1 like |
| chr12_-_56741535 | 0.08 |
ENST00000647707.1
|
ENSG00000285625.1
|
novel protein |
| chr20_-_37178966 | 0.08 |
ENST00000422138.1
|
MROH8
|
maestro heat like repeat family member 8 |
| chr11_+_74748831 | 0.08 |
ENST00000299563.5
|
RNF169
|
ring finger protein 169 |
| chr7_-_44541318 | 0.07 |
ENST00000381160.8
|
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr2_+_184598520 | 0.07 |
ENST00000302277.7
|
ZNF804A
|
zinc finger protein 804A |
| chrM_+_9207 | 0.07 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chr7_-_44541262 | 0.07 |
ENST00000289547.8
ENST00000546276.5 ENST00000423141.1 |
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr1_+_218285283 | 0.07 |
ENST00000366932.4
|
RRP15
|
ribosomal RNA processing 15 homolog |
| chr22_+_38705922 | 0.07 |
ENST00000216044.10
|
GTPBP1
|
GTP binding protein 1 |
| chr2_-_60553409 | 0.07 |
ENST00000358510.6
ENST00000643004.1 |
BCL11A
|
BAF chromatin remodeling complex subunit BCL11A |
| chr20_+_43558968 | 0.06 |
ENST00000647834.1
ENST00000373100.7 ENST00000648083.1 ENST00000648530.1 |
SGK2
|
serum/glucocorticoid regulated kinase 2 |
| chr4_+_73436198 | 0.06 |
ENST00000395792.7
|
AFP
|
alpha fetoprotein |
| chr4_+_73436244 | 0.06 |
ENST00000226359.2
|
AFP
|
alpha fetoprotein |
| chr14_+_39233908 | 0.05 |
ENST00000280082.4
|
MIA2
|
MIA SH3 domain ER export factor 2 |
| chr1_-_66801276 | 0.05 |
ENST00000304526.3
|
INSL5
|
insulin like 5 |
| chr3_-_157503375 | 0.05 |
ENST00000362010.7
|
VEPH1
|
ventricular zone expressed PH domain containing 1 |
| chr3_-_157503339 | 0.05 |
ENST00000392833.6
|
VEPH1
|
ventricular zone expressed PH domain containing 1 |
| chr7_-_7535863 | 0.05 |
ENST00000399429.8
|
COL28A1
|
collagen type XXVIII alpha 1 chain |
| chr19_-_35812838 | 0.05 |
ENST00000653904.2
|
PRODH2
|
proline dehydrogenase 2 |
| chr8_-_18684093 | 0.05 |
ENST00000428502.6
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr8_-_18684033 | 0.05 |
ENST00000614430.3
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr1_-_197067234 | 0.04 |
ENST00000367412.2
|
F13B
|
coagulation factor XIII B chain |
| chrX_-_123623155 | 0.04 |
ENST00000618150.4
|
THOC2
|
THO complex 2 |
| chrM_+_8489 | 0.04 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase membrane subunit 6 |
| chr3_-_49029378 | 0.04 |
ENST00000442157.2
ENST00000326739.9 ENST00000677010.1 ENST00000678724.1 ENST00000429182.6 |
IMPDH2
|
inosine monophosphate dehydrogenase 2 |
| chr15_+_58431985 | 0.04 |
ENST00000433326.2
ENST00000299022.10 |
LIPC
|
lipase C, hepatic type |
| chr1_-_46551647 | 0.04 |
ENST00000481882.7
|
KNCN
|
kinocilin |
| chr11_-_61920267 | 0.04 |
ENST00000531922.2
ENST00000301773.9 |
RAB3IL1
|
RAB3A interacting protein like 1 |
| chr12_+_130953898 | 0.04 |
ENST00000261654.10
|
ADGRD1
|
adhesion G protein-coupled receptor D1 |
| chr6_+_28349907 | 0.04 |
ENST00000252211.7
ENST00000341464.9 ENST00000377255.3 |
ZKSCAN3
|
zinc finger with KRAB and SCAN domains 3 |
| chr4_-_99435336 | 0.04 |
ENST00000437033.7
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr11_-_57324907 | 0.04 |
ENST00000358252.8
|
TNKS1BP1
|
tankyrase 1 binding protein 1 |
| chr7_+_151028422 | 0.04 |
ENST00000542328.5
ENST00000461373.5 ENST00000297504.10 ENST00000358849.9 ENST00000498578.5 ENST00000477719.5 ENST00000477092.5 |
ABCB8
|
ATP binding cassette subfamily B member 8 |
| chr1_-_159714581 | 0.04 |
ENST00000255030.9
ENST00000437342.1 ENST00000368112.5 ENST00000368111.5 ENST00000368110.1 |
CRP
|
C-reactive protein |
| chr17_+_7407838 | 0.04 |
ENST00000302926.7
|
NLGN2
|
neuroligin 2 |
| chr9_-_97938157 | 0.04 |
ENST00000616898.2
|
HEMGN
|
hemogen |
| chr22_-_30574572 | 0.04 |
ENST00000402369.5
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr1_-_160579439 | 0.04 |
ENST00000368054.8
ENST00000368048.7 ENST00000311224.8 ENST00000368051.3 ENST00000534968.5 |
CD84
|
CD84 molecule |
| chrX_-_18672101 | 0.03 |
ENST00000379984.4
|
RS1
|
retinoschisin 1 |
| chr4_-_53591477 | 0.03 |
ENST00000263925.8
|
LNX1
|
ligand of numb-protein X 1 |
| chr16_+_56191476 | 0.03 |
ENST00000262493.12
|
GNAO1
|
G protein subunit alpha o1 |
| chr2_-_60553618 | 0.03 |
ENST00000643716.1
ENST00000359629.10 ENST00000642384.2 ENST00000335712.11 |
BCL11A
|
BAF chromatin remodeling complex subunit BCL11A |
| chr3_+_35680994 | 0.03 |
ENST00000441454.5
|
ARPP21
|
cAMP regulated phosphoprotein 21 |
| chr12_-_55296569 | 0.03 |
ENST00000358433.3
|
OR6C6
|
olfactory receptor family 6 subfamily C member 6 |
| chr6_+_25754699 | 0.03 |
ENST00000439485.6
ENST00000377905.9 |
SLC17A4
|
solute carrier family 17 member 4 |
| chr6_+_160702238 | 0.03 |
ENST00000366924.6
ENST00000308192.14 ENST00000418964.1 |
PLG
|
plasminogen |
| chr19_+_53962925 | 0.03 |
ENST00000270458.4
|
CACNG8
|
calcium voltage-gated channel auxiliary subunit gamma 8 |
| chrX_-_15601077 | 0.03 |
ENST00000680121.1
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr1_+_159587817 | 0.03 |
ENST00000255040.3
|
APCS
|
amyloid P component, serum |
| chrY_+_18546691 | 0.03 |
ENST00000309834.8
ENST00000307393.3 ENST00000382856.2 |
HSFY1
|
heat shock transcription factor Y-linked 1 |
| chr6_-_30932147 | 0.02 |
ENST00000359086.4
|
SFTA2
|
surfactant associated 2 |
| chr19_-_45730861 | 0.02 |
ENST00000317683.4
|
FBXO46
|
F-box protein 46 |
| chr18_+_34976928 | 0.02 |
ENST00000591734.5
ENST00000413393.5 ENST00000589180.5 ENST00000587359.5 |
MAPRE2
|
microtubule associated protein RP/EB family member 2 |
| chr5_-_35195236 | 0.02 |
ENST00000509839.5
|
PRLR
|
prolactin receptor |
| chr2_+_132416795 | 0.02 |
ENST00000329321.4
|
GPR39
|
G protein-coupled receptor 39 |
| chr6_-_116126120 | 0.02 |
ENST00000452729.1
ENST00000651968.1 ENST00000243222.8 |
COL10A1
|
collagen type X alpha 1 chain |
| chr12_-_130716264 | 0.02 |
ENST00000643940.1
|
RIMBP2
|
RIMS binding protein 2 |
| chr7_+_48171451 | 0.02 |
ENST00000435803.6
|
ABCA13
|
ATP binding cassette subfamily A member 13 |
| chr3_-_165078480 | 0.02 |
ENST00000264382.8
|
SI
|
sucrase-isomaltase |
| chr6_+_47781982 | 0.02 |
ENST00000489301.6
ENST00000638973.1 ENST00000371211.6 ENST00000393699.2 |
OPN5
|
opsin 5 |
| chr3_-_167474026 | 0.02 |
ENST00000466903.1
ENST00000264677.8 |
SERPINI2
|
serpin family I member 2 |
| chr12_+_53954870 | 0.01 |
ENST00000243103.4
|
HOXC12
|
homeobox C12 |
| chr2_-_40453438 | 0.01 |
ENST00000455476.5
|
SLC8A1
|
solute carrier family 8 member A1 |
| chr2_+_233729042 | 0.01 |
ENST00000482026.6
|
UGT1A3
|
UDP glucuronosyltransferase family 1 member A3 |
| chr7_+_48924559 | 0.01 |
ENST00000650262.1
|
CDC14C
|
cell division cycle 14C |
| chr3_+_35680932 | 0.01 |
ENST00000396481.6
|
ARPP21
|
cAMP regulated phosphoprotein 21 |
| chr2_+_233718734 | 0.01 |
ENST00000373409.8
|
UGT1A4
|
UDP glucuronosyltransferase family 1 member A4 |
| chr6_-_29045175 | 0.01 |
ENST00000377175.2
|
OR2W1
|
olfactory receptor family 2 subfamily W member 1 |
| chr4_-_52020332 | 0.01 |
ENST00000682860.1
|
LRRC66
|
leucine rich repeat containing 66 |
| chr8_-_69833338 | 0.01 |
ENST00000524945.5
|
SLCO5A1
|
solute carrier organic anion transporter family member 5A1 |
| chr2_+_233712905 | 0.01 |
ENST00000373414.4
|
UGT1A5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr12_-_68159732 | 0.01 |
ENST00000229135.4
|
IFNG
|
interferon gamma |
| chr11_+_8019193 | 0.01 |
ENST00000534099.5
|
TUB
|
TUB bipartite transcription factor |
| chr6_-_32407123 | 0.01 |
ENST00000374993.4
ENST00000544175.2 ENST00000454136.7 ENST00000446536.2 |
BTNL2
|
butyrophilin like 2 |
| chr12_+_64497968 | 0.01 |
ENST00000676593.1
ENST00000677093.1 |
TBK1
ENSG00000288665.1
|
TANK binding kinase 1 novel transcript |
| chrX_+_85003863 | 0.00 |
ENST00000373173.7
|
APOOL
|
apolipoprotein O like |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:2000173 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.1 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.0 | 0.0 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.0 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030021 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.0 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |