Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
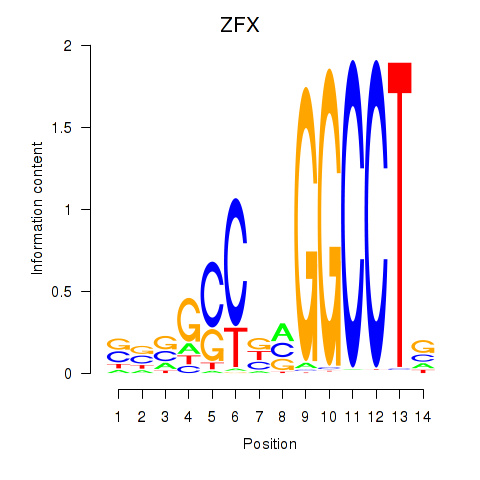
Results for ZFX
Z-value: 1.03
Transcription factors associated with ZFX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZFX
|
ENSG00000005889.16 | ZFX |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZFX | hg38_v1_chrX_+_24149699_24149773 | 0.15 | 4.8e-01 | Click! |
Activity profile of ZFX motif
Sorted Z-values of ZFX motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZFX
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_64049631 | 2.06 |
ENST00000340356.9
|
SOX18
|
SRY-box transcription factor 18 |
| chr1_+_61077219 | 1.95 |
ENST00000407417.7
|
NFIA
|
nuclear factor I A |
| chr20_+_43916142 | 1.82 |
ENST00000423191.6
ENST00000372999.5 |
TOX2
|
TOX high mobility group box family member 2 |
| chr9_+_126326809 | 1.40 |
ENST00000361171.8
ENST00000489637.3 |
MVB12B
|
multivesicular body subunit 12B |
| chr19_+_45340774 | 1.26 |
ENST00000589837.5
|
KLC3
|
kinesin light chain 3 |
| chr1_+_26111798 | 1.24 |
ENST00000374269.2
ENST00000374271.8 |
PDIK1L
|
PDLIM1 interacting kinase 1 like |
| chr9_+_35605277 | 1.21 |
ENST00000620767.4
|
TESK1
|
testis associated actin remodelling kinase 1 |
| chr9_+_35605234 | 1.19 |
ENST00000336395.6
|
TESK1
|
testis associated actin remodelling kinase 1 |
| chr4_+_41360759 | 1.18 |
ENST00000508501.5
ENST00000512946.5 ENST00000313860.11 ENST00000512632.5 ENST00000512820.5 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr19_-_17245889 | 1.18 |
ENST00000291442.4
|
NR2F6
|
nuclear receptor subfamily 2 group F member 6 |
| chr5_+_177133005 | 1.17 |
ENST00000510954.5
ENST00000354179.8 |
NSD1
|
nuclear receptor binding SET domain protein 1 |
| chr19_+_17226662 | 1.17 |
ENST00000598068.5
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr14_+_24398986 | 1.12 |
ENST00000382554.4
|
NYNRIN
|
NYN domain and retroviral integrase containing |
| chr2_-_86623807 | 1.10 |
ENST00000237455.5
|
RNF103
|
ring finger protein 103 |
| chr15_+_57376497 | 1.09 |
ENST00000281282.6
|
CGNL1
|
cingulin like 1 |
| chr10_+_117543567 | 1.08 |
ENST00000616794.1
|
EMX2
|
empty spiracles homeobox 2 |
| chr1_-_58577244 | 1.08 |
ENST00000371225.4
|
TACSTD2
|
tumor associated calcium signal transducer 2 |
| chr1_+_26111139 | 1.08 |
ENST00000619836.4
ENST00000444713.5 |
PDIK1L
|
PDLIM1 interacting kinase 1 like |
| chr9_+_137077467 | 1.06 |
ENST00000409858.8
|
UAP1L1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 like 1 |
| chr9_+_36036899 | 1.04 |
ENST00000377966.4
|
RECK
|
reversion inducing cysteine rich protein with kazal motifs |
| chr8_+_94895763 | 1.03 |
ENST00000523378.5
|
NDUFAF6
|
NADH:ubiquinone oxidoreductase complex assembly factor 6 |
| chr12_+_131894615 | 1.02 |
ENST00000321867.6
|
ULK1
|
unc-51 like autophagy activating kinase 1 |
| chr14_-_53956811 | 0.98 |
ENST00000559087.5
ENST00000245451.9 |
BMP4
|
bone morphogenetic protein 4 |
| chr7_+_90211830 | 0.93 |
ENST00000394622.6
ENST00000394632.5 ENST00000426158.1 ENST00000402625.6 |
STEAP2
|
STEAP2 metalloreductase |
| chr2_-_159286609 | 0.92 |
ENST00000409124.1
|
WDSUB1
|
WD repeat, sterile alpha motif and U-box domain containing 1 |
| chr7_+_90211686 | 0.92 |
ENST00000287908.7
ENST00000394621.7 ENST00000394626.5 |
STEAP2
|
STEAP2 metalloreductase |
| chr10_-_125158704 | 0.92 |
ENST00000531469.5
|
CTBP2
|
C-terminal binding protein 2 |
| chr4_+_155667654 | 0.91 |
ENST00000513574.1
|
GUCY1A1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr12_-_49828394 | 0.91 |
ENST00000335999.7
|
NCKAP5L
|
NCK associated protein 5 like |
| chr15_+_66293541 | 0.89 |
ENST00000319212.9
ENST00000525109.1 |
DIS3L
|
DIS3 like exosome 3'-5' exoribonuclease |
| chr19_+_17226597 | 0.87 |
ENST00000597836.5
|
OCEL1
|
occludin/ELL domain containing 1 |
| chr9_-_75088140 | 0.83 |
ENST00000361092.9
ENST00000376811.5 |
NMRK1
|
nicotinamide riboside kinase 1 |
| chr13_-_48533165 | 0.82 |
ENST00000430805.6
ENST00000544492.5 ENST00000544904.3 |
RCBTB2
|
RCC1 and BTB domain containing protein 2 |
| chr6_+_11537738 | 0.82 |
ENST00000379426.2
|
TMEM170B
|
transmembrane protein 170B |
| chr9_-_75088198 | 0.79 |
ENST00000376808.8
|
NMRK1
|
nicotinamide riboside kinase 1 |
| chr2_-_159286572 | 0.79 |
ENST00000409990.7
|
WDSUB1
|
WD repeat, sterile alpha motif and U-box domain containing 1 |
| chr2_-_159286546 | 0.79 |
ENST00000392796.7
|
WDSUB1
|
WD repeat, sterile alpha motif and U-box domain containing 1 |
| chr8_+_94895837 | 0.78 |
ENST00000519136.5
|
NDUFAF6
|
NADH:ubiquinone oxidoreductase complex assembly factor 6 |
| chr11_+_67056875 | 0.78 |
ENST00000532559.1
|
RHOD
|
ras homolog family member D |
| chr2_-_159286678 | 0.77 |
ENST00000359774.9
ENST00000358147.8 |
WDSUB1
|
WD repeat, sterile alpha motif and U-box domain containing 1 |
| chr17_-_42121330 | 0.75 |
ENST00000225916.10
|
KAT2A
|
lysine acetyltransferase 2A |
| chr16_-_2858154 | 0.75 |
ENST00000571228.1
ENST00000161006.8 |
PRSS22
|
serine protease 22 |
| chr7_+_17298642 | 0.74 |
ENST00000242057.9
|
AHR
|
aryl hydrocarbon receptor |
| chr8_-_143568854 | 0.74 |
ENST00000524906.5
ENST00000532862.1 ENST00000534459.5 |
MROH6
|
maestro heat like repeat family member 6 |
| chr3_+_134795248 | 0.73 |
ENST00000398015.8
|
EPHB1
|
EPH receptor B1 |
| chr12_-_56221701 | 0.73 |
ENST00000615206.4
ENST00000549038.5 ENST00000552244.5 |
RNF41
|
ring finger protein 41 |
| chr19_+_45497221 | 0.73 |
ENST00000456399.6
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent 1N (putative) |
| chr19_+_1354931 | 0.73 |
ENST00000591337.7
|
PWWP3A
|
PWWP domain containing 3A, DNA repair factor |
| chr20_+_36154630 | 0.73 |
ENST00000338074.7
ENST00000636016.2 ENST00000373945.5 |
EPB41L1
|
erythrocyte membrane protein band 4.1 like 1 |
| chr16_-_30096170 | 0.72 |
ENST00000566134.5
ENST00000565110.5 ENST00000398841.6 ENST00000398838.8 |
YPEL3
|
yippee like 3 |
| chr13_-_109786567 | 0.72 |
ENST00000375856.5
|
IRS2
|
insulin receptor substrate 2 |
| chr6_+_117482634 | 0.71 |
ENST00000296955.12
ENST00000338728.10 |
DCBLD1
|
discoidin, CUB and LCCL domain containing 1 |
| chr12_-_56221909 | 0.71 |
ENST00000394013.6
ENST00000345093.9 ENST00000551711.5 ENST00000552656.5 |
RNF41
|
ring finger protein 41 |
| chr5_+_42423433 | 0.70 |
ENST00000230882.9
|
GHR
|
growth hormone receptor |
| chr5_+_177133741 | 0.70 |
ENST00000439151.7
|
NSD1
|
nuclear receptor binding SET domain protein 1 |
| chr4_-_98657635 | 0.69 |
ENST00000515287.5
ENST00000511651.5 ENST00000505184.5 |
TSPAN5
|
tetraspanin 5 |
| chr19_+_50203607 | 0.69 |
ENST00000642316.2
ENST00000425460.6 ENST00000440075.6 ENST00000376970.6 ENST00000599920.5 |
MYH14
|
myosin heavy chain 14 |
| chr17_-_81911350 | 0.68 |
ENST00000570388.5
|
PCYT2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr19_+_17406099 | 0.68 |
ENST00000634942.2
ENST00000634568.1 ENST00000600514.5 |
BISPR
MVB12A
|
BST2 interferon stimulated positive regulator multivesicular body subunit 12A |
| chr22_+_50705495 | 0.68 |
ENST00000664402.1
|
SHANK3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr14_-_38256074 | 0.67 |
ENST00000342213.3
|
CLEC14A
|
C-type lectin domain containing 14A |
| chr1_+_215567279 | 0.66 |
ENST00000259154.9
|
KCTD3
|
potassium channel tetramerization domain containing 3 |
| chr17_+_49788672 | 0.66 |
ENST00000454930.6
ENST00000259021.9 ENST00000509773.5 ENST00000510819.5 ENST00000424009.6 |
KAT7
|
lysine acetyltransferase 7 |
| chr9_+_122941003 | 0.66 |
ENST00000373647.9
ENST00000402311.5 |
RABGAP1
|
RAB GTPase activating protein 1 |
| chr20_+_38805686 | 0.65 |
ENST00000299824.6
ENST00000373331.2 |
PPP1R16B
|
protein phosphatase 1 regulatory subunit 16B |
| chr9_+_127612222 | 0.65 |
ENST00000637953.1
ENST00000636962.2 |
STXBP1
|
syntaxin binding protein 1 |
| chr22_+_29307005 | 0.64 |
ENST00000406549.7
ENST00000610653.4 ENST00000621062.4 ENST00000611648.2 ENST00000618518.3 |
GAS2L1
|
growth arrest specific 2 like 1 |
| chr18_+_9334757 | 0.64 |
ENST00000262120.10
ENST00000581641.1 |
TWSG1
|
twisted gastrulation BMP signaling modulator 1 |
| chr11_+_67056805 | 0.64 |
ENST00000308831.7
|
RHOD
|
ras homolog family member D |
| chr20_+_59933761 | 0.64 |
ENST00000358293.7
|
FAM217B
|
family with sequence similarity 217 member B |
| chr9_+_127612257 | 0.63 |
ENST00000637173.2
ENST00000630492.2 ENST00000627871.2 ENST00000373302.8 ENST00000373299.5 ENST00000650920.1 ENST00000476182.3 |
STXBP1
|
syntaxin binding protein 1 |
| chr15_+_74812829 | 0.63 |
ENST00000309664.10
ENST00000379709.7 |
LMAN1L
|
lectin, mannose binding 1 like |
| chrX_-_136767322 | 0.63 |
ENST00000370620.5
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor 6 |
| chr6_-_145964330 | 0.63 |
ENST00000275233.12
ENST00000367505.6 |
SHPRH
|
SNF2 histone linker PHD RING helicase |
| chr6_-_118651522 | 0.63 |
ENST00000368491.8
|
CEP85L
|
centrosomal protein 85 like |
| chr19_+_45497246 | 0.62 |
ENST00000396737.6
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent 1N (putative) |
| chr1_-_53945661 | 0.62 |
ENST00000194214.10
|
HSPB11
|
heat shock protein family B (small) member 11 |
| chr19_+_17226218 | 0.62 |
ENST00000601529.5
ENST00000215061.9 ENST00000600232.5 |
OCEL1
|
occludin/ELL domain containing 1 |
| chr11_+_925809 | 0.62 |
ENST00000332231.9
|
AP2A2
|
adaptor related protein complex 2 subunit alpha 2 |
| chr1_+_84078043 | 0.61 |
ENST00000370689.6
ENST00000370688.7 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr12_+_8914525 | 0.61 |
ENST00000543824.5
|
PHC1
|
polyhomeotic homolog 1 |
| chr1_+_20633450 | 0.61 |
ENST00000321556.5
|
PINK1
|
PTEN induced kinase 1 |
| chr6_-_145964084 | 0.61 |
ENST00000438092.6
ENST00000629427.2 |
SHPRH
|
SNF2 histone linker PHD RING helicase |
| chr11_+_95089804 | 0.60 |
ENST00000278505.5
|
ENDOD1
|
endonuclease domain containing 1 |
| chr19_-_51372640 | 0.60 |
ENST00000600427.5
ENST00000221978.10 |
NKG7
|
natural killer cell granule protein 7 |
| chr9_-_13279407 | 0.60 |
ENST00000546205.5
|
MPDZ
|
multiple PDZ domain crumbs cell polarity complex component |
| chr10_+_22321056 | 0.60 |
ENST00000376663.8
|
BMI1
|
BMI1 proto-oncogene, polycomb ring finger |
| chr12_+_31959406 | 0.60 |
ENST00000540924.5
ENST00000312561.9 |
RESF1
|
retroelement silencing factor 1 |
| chr1_-_230978796 | 0.59 |
ENST00000522821.5
ENST00000366662.8 ENST00000366661.9 ENST00000522399.1 |
TTC13
|
tetratricopeptide repeat domain 13 |
| chr6_-_149864300 | 0.59 |
ENST00000239367.7
|
LRP11
|
LDL receptor related protein 11 |
| chr22_-_50261272 | 0.59 |
ENST00000395780.5
|
MAPK12
|
mitogen-activated protein kinase 12 |
| chr3_-_125375249 | 0.59 |
ENST00000484491.5
ENST00000492394.5 ENST00000471196.1 ENST00000468369.5 ENST00000485866.5 ENST00000360647.9 |
ZNF148
|
zinc finger protein 148 |
| chr2_-_130144994 | 0.59 |
ENST00000457413.1
ENST00000409128.5 ENST00000441670.1 ENST00000409234.3 ENST00000409943.8 ENST00000392984.7 ENST00000310463.10 |
CCDC74B
|
coiled-coil domain containing 74B |
| chr5_+_177022684 | 0.58 |
ENST00000506693.5
ENST00000358149.8 ENST00000512315.5 ENST00000503425.5 |
ZNF346
|
zinc finger protein 346 |
| chr2_+_24076817 | 0.58 |
ENST00000613899.4
|
FAM228B
|
family with sequence similarity 228 member B |
| chr17_-_16215520 | 0.58 |
ENST00000582357.5
ENST00000436828.5 ENST00000268712.8 ENST00000411510.5 ENST00000395857.7 |
NCOR1
|
nuclear receptor corepressor 1 |
| chr3_+_32106612 | 0.58 |
ENST00000282541.10
ENST00000425459.5 ENST00000431009.1 |
GPD1L
|
glycerol-3-phosphate dehydrogenase 1 like |
| chr17_+_60600178 | 0.58 |
ENST00000629650.2
ENST00000305921.8 |
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent 1D |
| chr6_-_31897200 | 0.58 |
ENST00000395728.7
ENST00000375528.8 |
EHMT2
|
euchromatic histone lysine methyltransferase 2 |
| chr3_+_49171590 | 0.57 |
ENST00000332780.4
|
KLHDC8B
|
kelch domain containing 8B |
| chr9_-_13279642 | 0.57 |
ENST00000319217.12
|
MPDZ
|
multiple PDZ domain crumbs cell polarity complex component |
| chr2_-_200510044 | 0.57 |
ENST00000359878.8
ENST00000409157.5 |
KCTD18
|
potassium channel tetramerization domain containing 18 |
| chr16_+_2969307 | 0.56 |
ENST00000576565.1
ENST00000318782.9 |
PAQR4
|
progestin and adipoQ receptor family member 4 |
| chr16_+_2969270 | 0.56 |
ENST00000293978.12
|
PAQR4
|
progestin and adipoQ receptor family member 4 |
| chr11_-_4608226 | 0.56 |
ENST00000300747.10
ENST00000526337.5 |
TRIM68
|
tripartite motif containing 68 |
| chr19_+_17470474 | 0.55 |
ENST00000598424.5
ENST00000252595.12 |
SLC27A1
|
solute carrier family 27 member 1 |
| chr1_-_40665435 | 0.55 |
ENST00000372683.1
|
RIMS3
|
regulating synaptic membrane exocytosis 3 |
| chr17_+_30970984 | 0.55 |
ENST00000443677.6
ENST00000324689.8 ENST00000328381.10 ENST00000535306.6 ENST00000580444.2 |
RNF135
|
ring finger protein 135 |
| chr11_+_67303497 | 0.55 |
ENST00000308127.9
|
SSH3
|
slingshot protein phosphatase 3 |
| chr4_+_89111521 | 0.55 |
ENST00000603357.3
|
TIGD2
|
tigger transposable element derived 2 |
| chr8_-_8386429 | 0.55 |
ENST00000615670.5
|
PRAG1
|
PEAK1 related, kinase-activating pseudokinase 1 |
| chr9_-_13279564 | 0.55 |
ENST00000541718.5
ENST00000447879.6 |
MPDZ
|
multiple PDZ domain crumbs cell polarity complex component |
| chr8_-_142777174 | 0.54 |
ENST00000652477.1
ENST00000614491.1 ENST00000613110.4 |
LYNX1
|
Ly6/neurotoxin 1 |
| chr2_-_85418421 | 0.54 |
ENST00000409275.1
|
CAPG
|
capping actin protein, gelsolin like |
| chr11_-_6320494 | 0.54 |
ENST00000303927.4
ENST00000530979.1 |
CAVIN3
|
caveolae associated protein 3 |
| chr17_-_66192125 | 0.54 |
ENST00000535342.7
|
CEP112
|
centrosomal protein 112 |
| chr9_+_136712531 | 0.53 |
ENST00000371692.9
|
DIPK1B
|
divergent protein kinase domain 1B |
| chr10_-_50623897 | 0.53 |
ENST00000361781.7
ENST00000429490.5 ENST00000619438.4 |
SGMS1
|
sphingomyelin synthase 1 |
| chr4_-_109729987 | 0.53 |
ENST00000243501.10
|
PLA2G12A
|
phospholipase A2 group XIIA |
| chr11_+_925840 | 0.53 |
ENST00000448903.7
ENST00000525796.5 ENST00000534328.5 |
AP2A2
|
adaptor related protein complex 2 subunit alpha 2 |
| chr11_+_67303476 | 0.53 |
ENST00000376757.9
ENST00000308298.11 |
SSH3
|
slingshot protein phosphatase 3 |
| chr2_+_86913812 | 0.53 |
ENST00000398193.8
|
RGPD1
|
RANBP2 like and GRIP domain containing 1 |
| chr10_-_124093582 | 0.52 |
ENST00000462406.1
ENST00000435907.6 |
CHST15
|
carbohydrate sulfotransferase 15 |
| chr1_-_227947924 | 0.52 |
ENST00000272164.6
|
WNT9A
|
Wnt family member 9A |
| chr4_+_88007624 | 0.52 |
ENST00000237596.7
|
PKD2
|
polycystin 2, transient receptor potential cation channel |
| chr6_-_111483700 | 0.52 |
ENST00000435970.5
ENST00000358835.7 |
REV3L
|
REV3 like, DNA directed polymerase zeta catalytic subunit |
| chr14_+_66508495 | 0.52 |
ENST00000543237.5
|
GPHN
|
gephyrin |
| chr22_-_19178402 | 0.52 |
ENST00000451283.5
|
SLC25A1
|
solute carrier family 25 member 1 |
| chr2_+_169694434 | 0.52 |
ENST00000616481.4
ENST00000616524.4 ENST00000617738.4 ENST00000359744.8 ENST00000438838.5 ENST00000438710.5 ENST00000449906.5 ENST00000498202.6 ENST00000272797.8 |
PHOSPHO2
KLHL23
|
phosphatase, orphan 2 kelch like family member 23 |
| chr12_+_125065427 | 0.52 |
ENST00000316519.11
|
AACS
|
acetoacetyl-CoA synthetase |
| chr19_-_1812194 | 0.51 |
ENST00000525591.5
|
ATP8B3
|
ATPase phospholipid transporting 8B3 |
| chr19_+_35000275 | 0.51 |
ENST00000317991.10
ENST00000680623.1 |
GRAMD1A
|
GRAM domain containing 1A |
| chr7_+_100612372 | 0.51 |
ENST00000493970.5
|
MOSPD3
|
motile sperm domain containing 3 |
| chr1_-_16352420 | 0.50 |
ENST00000375592.8
|
FBXO42
|
F-box protein 42 |
| chr19_+_35000426 | 0.50 |
ENST00000411896.6
ENST00000424536.2 |
GRAMD1A
|
GRAM domain containing 1A |
| chr19_+_32405758 | 0.50 |
ENST00000392250.7
|
DPY19L3
|
dpy-19 like C-mannosyltransferase 3 |
| chr17_+_81035122 | 0.50 |
ENST00000321280.11
ENST00000575989.5 ENST00000428708.7 ENST00000575712.5 ENST00000575245.5 ENST00000435091.7 ENST00000321300.10 |
BAIAP2
|
BAR/IMD domain containing adaptor protein 2 |
| chr7_-_149773548 | 0.50 |
ENST00000302017.4
|
ZNF467
|
zinc finger protein 467 |
| chr15_+_42575592 | 0.50 |
ENST00000290607.12
|
STARD9
|
StAR related lipid transfer domain containing 9 |
| chr16_+_2471284 | 0.49 |
ENST00000293973.2
|
NTN3
|
netrin 3 |
| chr9_+_69325168 | 0.49 |
ENST00000303068.14
|
FAM189A2
|
family with sequence similarity 189 member A2 |
| chr19_-_11197516 | 0.49 |
ENST00000592903.5
ENST00000586659.6 ENST00000589359.5 ENST00000588724.5 |
KANK2
|
KN motif and ankyrin repeat domains 2 |
| chr7_-_15686671 | 0.49 |
ENST00000262041.6
|
MEOX2
|
mesenchyme homeobox 2 |
| chr18_-_812230 | 0.48 |
ENST00000314574.5
|
YES1
|
YES proto-oncogene 1, Src family tyrosine kinase |
| chr19_+_32406076 | 0.48 |
ENST00000342179.9
ENST00000586427.1 |
DPY19L3
|
dpy-19 like C-mannosyltransferase 3 |
| chr8_-_144465343 | 0.48 |
ENST00000526887.5
ENST00000533764.5 ENST00000403000.6 ENST00000306145.10 |
CYHR1
|
cysteine and histidine rich 1 |
| chr13_+_20703677 | 0.48 |
ENST00000682841.1
|
IL17D
|
interleukin 17D |
| chr7_-_149773207 | 0.48 |
ENST00000484747.5
|
ZNF467
|
zinc finger protein 467 |
| chr5_+_177022728 | 0.48 |
ENST00000511834.5
ENST00000503039.1 |
ZNF346
|
zinc finger protein 346 |
| chr9_-_5833014 | 0.48 |
ENST00000339450.10
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr17_-_66191855 | 0.48 |
ENST00000583358.1
ENST00000392769.6 |
CEP112
|
centrosomal protein 112 |
| chr9_+_2017383 | 0.48 |
ENST00000382194.6
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr5_-_149551381 | 0.48 |
ENST00000670598.1
ENST00000657001.1 ENST00000515768.6 ENST00000261798.10 ENST00000377843.8 |
CSNK1A1
|
casein kinase 1 alpha 1 |
| chr19_+_48624952 | 0.48 |
ENST00000599748.5
ENST00000599029.2 |
SPHK2
|
sphingosine kinase 2 |
| chr15_+_40323683 | 0.47 |
ENST00000638170.2
|
INAFM2
|
InaF motif containing 2 |
| chr5_-_14871757 | 0.47 |
ENST00000284268.8
|
ANKH
|
ANKH inorganic pyrophosphate transport regulator |
| chr13_-_48533069 | 0.47 |
ENST00000344532.8
|
RCBTB2
|
RCC1 and BTB domain containing protein 2 |
| chr22_-_19178719 | 0.47 |
ENST00000215882.10
|
SLC25A1
|
solute carrier family 25 member 1 |
| chr9_-_34376878 | 0.47 |
ENST00000297625.8
|
MYORG
|
myogenesis regulating glycosidase (putative) |
| chr1_+_25543598 | 0.47 |
ENST00000374338.5
|
LDLRAP1
|
low density lipoprotein receptor adaptor protein 1 |
| chr2_-_240682879 | 0.47 |
ENST00000407834.4
ENST00000621682.4 |
AQP12B
|
aquaporin 12B |
| chr17_-_37359076 | 0.46 |
ENST00000621960.1
ENST00000614428.4 ENST00000614789.4 |
ACACA
|
acetyl-CoA carboxylase alpha |
| chr15_+_56918763 | 0.46 |
ENST00000557843.5
|
TCF12
|
transcription factor 12 |
| chr8_+_94895813 | 0.46 |
ENST00000396113.5
|
NDUFAF6
|
NADH:ubiquinone oxidoreductase complex assembly factor 6 |
| chr3_+_12796662 | 0.46 |
ENST00000456430.6
ENST00000626378.1 |
CAND2
|
cullin associated and neddylation dissociated 2 (putative) |
| chr12_+_8914464 | 0.46 |
ENST00000544916.6
|
PHC1
|
polyhomeotic homolog 1 |
| chr8_-_142777802 | 0.46 |
ENST00000621401.4
|
LYNX1
|
Ly6/neurotoxin 1 |
| chr1_+_52726441 | 0.46 |
ENST00000294353.7
ENST00000545132.5 |
ZYG11B
|
zyg-11 family member B, cell cycle regulator |
| chr15_+_84817346 | 0.46 |
ENST00000258888.6
|
ALPK3
|
alpha kinase 3 |
| chr2_-_87825767 | 0.46 |
ENST00000638730.1
|
RGPD2
|
RANBP2 like and GRIP domain containing 2 |
| chr2_-_165794190 | 0.45 |
ENST00000392701.8
ENST00000422973.1 |
GALNT3
|
polypeptide N-acetylgalactosaminyltransferase 3 |
| chr2_+_131527833 | 0.45 |
ENST00000295171.10
ENST00000467992.6 ENST00000409856.8 |
CCDC74A
|
coiled-coil domain containing 74A |
| chr17_+_45160694 | 0.45 |
ENST00000307275.7
ENST00000585340.2 |
HEXIM2
|
HEXIM P-TEFb complex subunit 2 |
| chr2_+_188292771 | 0.45 |
ENST00000359135.7
|
GULP1
|
GULP PTB domain containing engulfment adaptor 1 |
| chr12_+_66189208 | 0.45 |
ENST00000261233.9
|
IRAK3
|
interleukin 1 receptor associated kinase 3 |
| chr19_+_2096960 | 0.45 |
ENST00000588003.5
|
IZUMO4
|
IZUMO family member 4 |
| chr12_+_48106094 | 0.45 |
ENST00000546755.5
ENST00000549366.5 ENST00000642730.1 ENST00000552792.5 |
PFKM
|
phosphofructokinase, muscle |
| chr17_-_5584448 | 0.44 |
ENST00000269280.8
ENST00000571451.6 ENST00000572272.6 ENST00000613500.4 ENST00000619223.4 ENST00000617618.4 ENST00000345221.7 ENST00000262467.10 |
NLRP1
|
NLR family pyrin domain containing 1 |
| chr13_-_110915006 | 0.44 |
ENST00000310847.8
ENST00000267339.6 |
ANKRD10
|
ankyrin repeat domain 10 |
| chr20_+_11890785 | 0.44 |
ENST00000399006.6
ENST00000405977.5 |
BTBD3
|
BTB domain containing 3 |
| chr5_+_113513674 | 0.44 |
ENST00000161863.9
ENST00000515883.5 |
YTHDC2
|
YTH domain containing 2 |
| chr7_+_91940970 | 0.44 |
ENST00000359028.7
|
AKAP9
|
A-kinase anchoring protein 9 |
| chr11_-_47185840 | 0.44 |
ENST00000539589.5
ENST00000528462.5 |
PACSIN3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr1_-_100132892 | 0.44 |
ENST00000287482.6
|
SASS6
|
SAS-6 centriolar assembly protein |
| chr20_+_64255728 | 0.44 |
ENST00000369758.8
ENST00000308824.11 ENST00000609372.1 ENST00000610196.1 ENST00000609764.1 |
PCMTD2
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
| chr2_+_172735838 | 0.44 |
ENST00000397081.8
|
RAPGEF4
|
Rap guanine nucleotide exchange factor 4 |
| chr9_+_135479772 | 0.44 |
ENST00000356818.7
|
PPP1R26
|
protein phosphatase 1 regulatory subunit 26 |
| chr19_+_32405789 | 0.44 |
ENST00000586987.5
|
DPY19L3
|
dpy-19 like C-mannosyltransferase 3 |
| chr16_+_2969548 | 0.43 |
ENST00000572687.1
|
PAQR4
|
progestin and adipoQ receptor family member 4 |
| chr20_-_25585517 | 0.43 |
ENST00000422516.5
ENST00000278886.11 |
NINL
|
ninein like |
| chr14_-_35809248 | 0.43 |
ENST00000554259.6
ENST00000680220.1 ENST00000307138.10 |
RALGAPA1
|
Ral GTPase activating protein catalytic subunit alpha 1 |
| chr20_+_11890723 | 0.43 |
ENST00000254977.7
|
BTBD3
|
BTB domain containing 3 |
| chr2_+_240691851 | 0.43 |
ENST00000429564.1
ENST00000337801.9 |
AQP12A
|
aquaporin 12A |
| chr7_-_36367141 | 0.43 |
ENST00000453212.5
ENST00000415803.2 ENST00000431396.1 ENST00000440378.6 ENST00000317020.10 ENST00000436884.5 |
KIAA0895
|
KIAA0895 |
| chr2_-_39437264 | 0.43 |
ENST00000263881.8
ENST00000341681.9 |
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr1_-_53945567 | 0.43 |
ENST00000371378.6
|
HSPB11
|
heat shock protein family B (small) member 11 |
| chr17_-_81911128 | 0.43 |
ENST00000573927.5
ENST00000331285.7 ENST00000572157.1 |
PCYT2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr14_-_35809206 | 0.43 |
ENST00000637992.1
ENST00000389698.7 |
RALGAPA1
|
Ral GTPase activating protein catalytic subunit alpha 1 |
| chr22_+_19714450 | 0.42 |
ENST00000455784.7
ENST00000406395.5 |
SEPTIN5
|
septin 5 |
| chr12_+_103930600 | 0.42 |
ENST00000680316.1
ENST00000679861.1 |
HSP90B1
|
heat shock protein 90 beta family member 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | GO:0019075 | virus maturation(GO:0019075) |
| 0.4 | 2.4 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.4 | 1.6 | GO:1903296 | regulation of glutamate secretion, neurotransmission(GO:1903294) positive regulation of glutamate secretion, neurotransmission(GO:1903296) |
| 0.4 | 1.1 | GO:0099557 | trans-synaptic signaling by trans-synaptic complex, modulating synaptic transmission(GO:0099557) |
| 0.3 | 1.0 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.3 | 1.0 | GO:0072096 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.3 | 0.9 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.3 | 0.8 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.3 | 1.1 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.3 | 0.5 | GO:0043465 | fermentation(GO:0006113) regulation of fermentation(GO:0043465) |
| 0.3 | 1.1 | GO:0035261 | external genitalia morphogenesis(GO:0035261) |
| 0.3 | 1.8 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.3 | 0.8 | GO:0042040 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.3 | 1.8 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.2 | 0.7 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.2 | 0.6 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.2 | 0.6 | GO:1903383 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) |
| 0.2 | 0.6 | GO:0010931 | macrophage tolerance induction(GO:0010931) regulation of macrophage tolerance induction(GO:0010932) positive regulation of macrophage tolerance induction(GO:0010933) |
| 0.2 | 0.6 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.2 | 0.6 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 | 0.6 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.2 | 0.6 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.2 | 0.6 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.2 | 0.5 | GO:0046707 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.2 | 0.5 | GO:0071464 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) cellular response to hydrostatic pressure(GO:0071464) |
| 0.2 | 0.7 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.2 | 0.5 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.2 | 0.5 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.2 | 0.5 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.2 | 0.8 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 1.0 | GO:0075071 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.1 | 0.4 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.1 | 0.7 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.1 | 1.4 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.1 | 0.4 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.1 | 0.7 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 | 0.4 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.1 | 1.3 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 | 0.8 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.1 | 1.2 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.1 | 0.4 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.1 | 0.4 | GO:0061445 | endocardial cell fate commitment(GO:0060957) endocardial cushion cell fate commitment(GO:0061445) |
| 0.1 | 0.4 | GO:0061010 | gall bladder development(GO:0061010) hepatic duct development(GO:0061011) |
| 0.1 | 0.5 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.1 | 0.7 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 0.4 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.1 | 0.3 | GO:2000176 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.1 | 0.3 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.1 | 0.6 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.4 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.1 | 0.3 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.8 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 0.5 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.6 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.1 | 0.6 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.3 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.1 | 0.4 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 0.5 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.1 | 0.3 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.1 | 0.4 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.1 | 0.3 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.1 | 0.2 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.1 | 0.3 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 0.4 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.4 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.1 | 0.4 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.1 | 0.6 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.3 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.1 | 0.5 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.1 | 0.3 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.1 | 0.8 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 | 0.6 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.4 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.1 | 0.9 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.5 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.1 | 0.6 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 0.7 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 0.1 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.1 | 0.2 | GO:0071206 | establishment of protein localization to juxtaparanode region of axon(GO:0071206) |
| 0.1 | 0.2 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.1 | 1.0 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 1.9 | GO:0072189 | ureter development(GO:0072189) |
| 0.1 | 0.2 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.2 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.1 | 1.3 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 | 0.3 | GO:0035854 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) regulation of forebrain neuron differentiation(GO:2000977) |
| 0.1 | 0.6 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.1 | 0.4 | GO:1901963 | regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) |
| 0.1 | 0.4 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.1 | 0.6 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.2 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 | 0.4 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 | 0.1 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.1 | 0.6 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.1 | 0.4 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.6 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.2 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 0.7 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.1 | 0.2 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.1 | 0.5 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 1.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.2 | GO:0031585 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) |
| 0.1 | 0.2 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 0.3 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.2 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.3 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 0.8 | GO:0036481 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036481) |
| 0.1 | 0.9 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.2 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.1 | 0.2 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 0.7 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 0.2 | GO:0099403 | negative regulation of maintenance of sister chromatid cohesion(GO:0034092) negative regulation of maintenance of mitotic sister chromatid cohesion(GO:0034183) maintenance of mitotic sister chromatid cohesion, telomeric(GO:0099403) mitotic sister chromatid cohesion, telomeric(GO:0099404) regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904907) negative regulation of maintenance of mitotic sister chromatid cohesion, telomeric(GO:1904908) |
| 0.1 | 1.5 | GO:0046337 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.1 | 0.6 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.1 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.1 | 0.4 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.2 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.1 | 0.2 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.1 | 0.4 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.1 | 0.4 | GO:0036486 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.1 | 0.2 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.1 | 0.5 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.2 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 | 0.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 1.4 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.1 | 0.2 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.1 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.8 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.1 | 0.2 | GO:0060382 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 0.1 | 0.2 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.1 | 1.4 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.6 | GO:0007379 | segment specification(GO:0007379) |
| 0.1 | 0.2 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 0.1 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.1 | 0.2 | GO:0071373 | cellular response to cocaine(GO:0071314) cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.1 | 0.2 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.8 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 0.2 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.1 | 0.2 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.1 | 0.4 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.3 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 0.2 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.1 | 0.2 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.2 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.3 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.3 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.1 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.7 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.2 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.6 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.9 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.2 | GO:0006404 | RNA import into nucleus(GO:0006404) |
| 0.0 | 0.2 | GO:0002741 | positive regulation of cytokine secretion involved in immune response(GO:0002741) |
| 0.0 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.6 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.3 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.2 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.2 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.0 | 0.9 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.2 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.0 | 0.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.6 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.3 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:0036404 | conversion of ds siRNA to ss siRNA involved in RNA interference(GO:0033168) conversion of ds siRNA to ss siRNA(GO:0036404) |
| 0.0 | 0.4 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.7 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.5 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.4 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.3 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.2 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 0.3 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.1 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.0 | 0.4 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.5 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.2 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.1 | GO:1904395 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 0.2 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.1 | GO:0019413 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.0 | 0.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0006524 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) L-alanine metabolic process(GO:0042851) L-alanine catabolic process(GO:0042853) |
| 0.0 | 0.1 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.2 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.0 | 0.3 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.2 | GO:0061084 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.3 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.2 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.3 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.6 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.4 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.4 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.4 | GO:1902514 | regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 1.4 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.1 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 1.4 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.6 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.8 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.3 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.0 | 0.2 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.0 | 0.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 1.0 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.5 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.2 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.0 | 0.2 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.2 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.0 | GO:1900222 | negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.0 | 0.2 | GO:0044858 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.8 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.2 | GO:0019086 | late viral transcription(GO:0019086) |
| 0.0 | 0.1 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.5 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.0 | 0.0 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.2 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.2 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.2 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.1 | GO:0070676 | intralumenal vesicle formation(GO:0070676) negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 | 0.2 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.5 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.0 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.1 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0090034 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.0 | 0.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.1 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.7 | GO:0036120 | response to platelet-derived growth factor(GO:0036119) cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.1 | GO:0003064 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.3 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.5 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.2 | GO:0007549 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.3 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0072312 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.0 | 0.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0033085 | negative regulation of T cell differentiation in thymus(GO:0033085) negative regulation of thymocyte aggregation(GO:2000399) |
| 0.0 | 0.6 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.7 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.7 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.0 | 0.4 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 1.5 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.3 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.6 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 1.2 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.1 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) mammary duct terminal end bud growth(GO:0060763) |
| 0.0 | 0.3 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.2 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.2 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.4 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.1 | GO:2000374 | regulation of oxygen metabolic process(GO:2000374) |
| 0.0 | 0.1 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.1 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.0 | 0.6 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.2 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 1.1 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.1 | GO:1903939 | regulation of TORC2 signaling(GO:1903939) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.3 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.3 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.1 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 0.2 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.0 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.0 | 0.3 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 1.8 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.7 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.0 | 0.7 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.6 | GO:0009251 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.4 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0072710 | response to hydroxyurea(GO:0072710) |
| 0.0 | 0.8 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.2 | GO:0090179 | regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.1 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.6 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.0 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.4 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.1 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 0.0 | 0.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 1.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.1 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.0 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.4 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.2 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.1 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.4 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.2 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.4 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.2 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.6 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.7 | GO:0098534 | centriole assembly(GO:0098534) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.6 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.3 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.0 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.0 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.0 | 0.1 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.0 | 0.3 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.1 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.1 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.1 | GO:1990166 | positive regulation of DNA ligation(GO:0051106) protein localization to site of double-strand break(GO:1990166) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.0 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.0 | 0.3 | GO:0071467 | cellular response to pH(GO:0071467) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.3 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.3 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.1 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.0 | 0.4 | GO:0097502 | mannosylation(GO:0097502) |
| 0.0 | 0.3 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 1.3 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.9 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.0 | GO:0042182 | ketone catabolic process(GO:0042182) |
| 0.0 | 0.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.1 | GO:0021903 | rostrocaudal neural tube patterning(GO:0021903) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.2 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.2 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.0 | 0.6 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.2 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.4 | GO:0097503 | sialylation(GO:0097503) |
| 0.0 | 0.5 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.2 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.0 | GO:0003290 | septum secundum development(GO:0003285) atrial septum secundum morphogenesis(GO:0003290) |
| 0.0 | 0.3 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.2 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.0 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.9 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.6 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.3 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.3 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.0 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.3 | 1.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 0.7 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.2 | 0.7 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.2 | 0.9 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.2 | 2.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 0.6 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 0.4 | GO:0032116 | SMC loading complex(GO:0032116) |
| 0.1 | 0.8 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.1 | 0.8 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 2.6 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 1.9 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.5 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.4 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.1 | 0.3 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 0.5 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.7 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 1.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.8 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.3 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.1 | 0.3 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.1 | 0.3 | GO:0097196 | Shu complex(GO:0097196) |
| 0.1 | 0.2 | GO:0070701 | mucus layer(GO:0070701) |
| 0.1 | 0.3 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.1 | 0.2 | GO:0097545 | axonemal outer doublet(GO:0097545) |
| 0.1 | 0.2 | GO:1905103 | integral component of lysosomal membrane(GO:1905103) |
| 0.1 | 0.4 | GO:0032449 | CBM complex(GO:0032449) |
| 0.1 | 1.4 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 0.5 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.3 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.1 | 0.1 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.1 | 2.1 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 0.6 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.7 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 1.1 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.9 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 0.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.4 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.6 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.6 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.2 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.2 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.2 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.0 | 0.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.7 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 1.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0033167 | ARC complex(GO:0033167) |
| 0.0 | 0.3 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 1.1 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.2 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.2 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.1 | GO:0000805 | X chromosome(GO:0000805) |
| 0.0 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.2 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.2 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.8 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0034677 | integrin alpha7-beta1 complex(GO:0034677) |
| 0.0 | 0.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.1 | GO:0000229 | cytoplasmic chromosome(GO:0000229) |
| 0.0 | 0.3 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.5 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 1.0 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.8 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 1.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.2 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.0 | 0.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 1.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.0 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0071756 | IgM immunoglobulin complex(GO:0071753) IgM immunoglobulin complex, circulating(GO:0071754) pentameric IgM immunoglobulin complex(GO:0071756) |
| 0.0 | 0.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.6 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.1 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.0 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.6 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 1.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 2.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.0 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0005128 | erythropoietin receptor binding(GO:0005128) interleukin-3 receptor binding(GO:0005135) |
| 0.4 | 1.6 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.4 | 1.5 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.3 | 1.8 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.3 | 0.8 | GO:0061599 | nitrate reductase activity(GO:0008940) molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.2 | 1.2 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.2 | 0.7 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.2 | 0.6 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.2 | 0.6 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.2 | 1.6 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.2 | 0.6 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.2 | 1.9 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.2 | 0.6 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.2 | 0.5 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.2 | 1.0 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.2 | 0.5 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.2 | 0.6 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.4 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 1.0 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.7 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.5 | GO:0019144 | ADP-sugar diphosphatase activity(GO:0019144) |
| 0.1 | 0.5 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 0.8 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 0.4 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.1 | 0.6 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.4 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.1 | 0.3 | GO:0047750 | cholestenol delta-isomerase activity(GO:0047750) |
| 0.1 | 0.5 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.5 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.1 | 1.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.1 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.1 | 0.6 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 1.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 0.6 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.1 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 1.0 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.4 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 0.3 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.1 | 0.3 | GO:0004314 | [acyl-carrier-protein] S-malonyltransferase activity(GO:0004314) 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) S-malonyltransferase activity(GO:0016419) malonyltransferase activity(GO:0016420) |
| 0.1 | 0.5 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.6 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 0.4 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 0.4 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.4 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.5 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.1 | 0.5 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 0.8 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 1.0 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.2 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.1 | 0.3 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.3 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.1 | 0.9 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 1.7 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 0.7 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 0.3 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.1 | 1.9 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 0.3 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 1.8 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 0.5 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 1.0 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.2 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.3 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.1 | 0.2 | GO:0051800 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.1 | 0.2 | GO:0000252 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) sterol-4-alpha-carboxylate 3-dehydrogenase (decarboxylating) activity(GO:0047012) |
| 0.1 | 0.2 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.1 | 0.4 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 0.3 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 1.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.7 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 0.6 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.7 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.6 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.2 | GO:0047661 | L-serine ammonia-lyase activity(GO:0003941) racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.1 | 0.5 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.2 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 0.3 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.1 | 0.3 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.1 | 0.7 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 0.6 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 0.7 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.4 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.1 | 0.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.2 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.1 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.5 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.1 | 0.4 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.1 | 0.4 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.9 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.2 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.0 | 0.2 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.2 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.0 | 0.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.7 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 1.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.1 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.0 | 0.3 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.5 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.6 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0031762 | alpha-1A adrenergic receptor binding(GO:0031691) follicle-stimulating hormone receptor binding(GO:0031762) |
| 0.0 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.0 | 0.2 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 1.0 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0052835 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol pentakisphosphate phosphatase activity(GO:0052827) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.0 | 1.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.4 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 1.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.2 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 1.0 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 1.4 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 1.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.6 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0070704 | C-5 sterol desaturase activity(GO:0000248) sterol desaturase activity(GO:0070704) |
| 0.0 | 1.1 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) calcitonin receptor binding(GO:0031716) |
| 0.0 | 0.1 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.2 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.2 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.2 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.4 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.4 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 1.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 1.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.3 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.5 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.2 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.8 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.2 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.3 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.3 | GO:0010857 | calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.7 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.5 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.1 | GO:0016497 | substance K receptor activity(GO:0016497) |
| 0.0 | 3.5 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.6 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.7 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) annealing activity(GO:0097617) |
| 0.0 | 0.6 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 1.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.9 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.0 | 0.3 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.0 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.4 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.4 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.0 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0098634 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.7 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.2 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.4 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.2 | 0.5 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 1.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 0.9 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.0 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 2.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 2.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.2 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.7 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 1.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.6 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.2 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.7 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.1 | 3.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.6 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 1.8 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 1.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 0.9 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 0.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.9 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.0 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.8 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 1.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 1.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 1.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.6 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.1 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 1.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.9 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.0 | 0.7 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.7 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.8 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.4 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.3 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.9 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.1 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |