Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
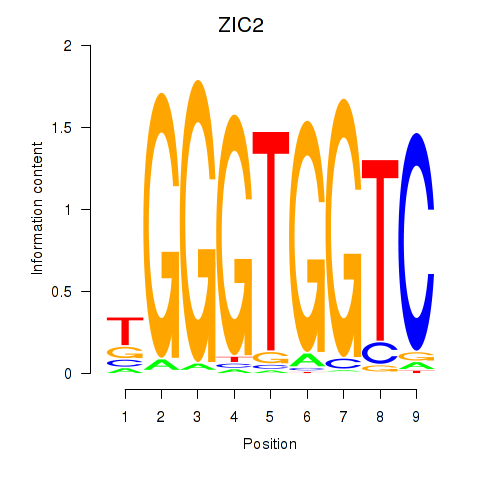
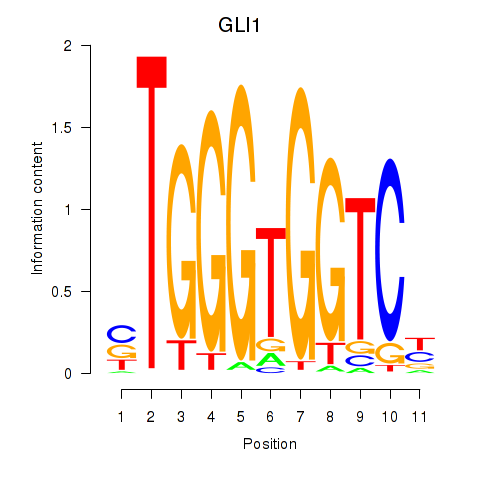
Results for ZIC2_GLI1
Z-value: 0.92
Transcription factors associated with ZIC2_GLI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZIC2
|
ENSG00000043355.12 | ZIC2 |
|
GLI1
|
ENSG00000111087.10 | GLI1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZIC2 | hg38_v1_chr13_+_99981775_99981791 | -0.14 | 5.1e-01 | Click! |
| GLI1 | hg38_v1_chr12_+_57459782_57459796, hg38_v1_chr12_+_57460127_57460151 | -0.07 | 7.5e-01 | Click! |
Activity profile of ZIC2_GLI1 motif
Sorted Z-values of ZIC2_GLI1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZIC2_GLI1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_205813177 | 5.28 |
ENST00000367137.4
|
SLC41A1
|
solute carrier family 41 member 1 |
| chr1_-_44843240 | 2.72 |
ENST00000372192.4
|
PTCH2
|
patched 2 |
| chr11_-_111912871 | 2.58 |
ENST00000528628.5
|
CRYAB
|
crystallin alpha B |
| chr4_-_5019437 | 2.46 |
ENST00000506508.1
ENST00000509419.1 ENST00000307746.9 |
CYTL1
|
cytokine like 1 |
| chr17_+_42682470 | 1.71 |
ENST00000264638.9
|
CNTNAP1
|
contactin associated protein 1 |
| chr21_-_26843012 | 1.64 |
ENST00000517777.6
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr21_-_26843063 | 1.58 |
ENST00000678221.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr8_+_31639755 | 1.55 |
ENST00000520407.5
|
NRG1
|
neuregulin 1 |
| chr10_-_88952763 | 1.55 |
ENST00000224784.10
|
ACTA2
|
actin alpha 2, smooth muscle |
| chr12_-_57129001 | 1.55 |
ENST00000556155.5
|
STAT6
|
signal transducer and activator of transcription 6 |
| chr6_-_32853618 | 1.51 |
ENST00000354258.5
|
TAP1
|
transporter 1, ATP binding cassette subfamily B member |
| chr6_-_32853813 | 1.49 |
ENST00000643049.2
|
TAP1
|
transporter 1, ATP binding cassette subfamily B member |
| chr6_-_29628038 | 1.42 |
ENST00000355973.7
ENST00000377012.8 |
GABBR1
|
gamma-aminobutyric acid type B receptor subunit 1 |
| chr5_+_162067764 | 1.33 |
ENST00000639213.2
ENST00000414552.6 |
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr5_+_162067858 | 1.32 |
ENST00000361925.9
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr8_+_31639222 | 1.23 |
ENST00000519301.6
ENST00000652698.1 |
NRG1
|
neuregulin 1 |
| chr8_+_31639291 | 1.23 |
ENST00000651149.1
ENST00000650866.1 |
NRG1
|
neuregulin 1 |
| chr5_+_162067990 | 1.22 |
ENST00000641017.1
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr5_+_162068031 | 1.20 |
ENST00000356592.8
|
GABRG2
|
gamma-aminobutyric acid type A receptor subunit gamma2 |
| chr9_+_106863121 | 1.16 |
ENST00000472574.1
ENST00000277225.10 |
ZNF462
|
zinc finger protein 462 |
| chr7_-_92836555 | 1.08 |
ENST00000424848.3
|
CDK6
|
cyclin dependent kinase 6 |
| chr7_-_108456321 | 1.03 |
ENST00000379024.8
ENST00000351718.8 |
NRCAM
|
neuronal cell adhesion molecule |
| chr16_+_75222609 | 1.02 |
ENST00000495583.1
|
CTRB1
|
chymotrypsinogen B1 |
| chr1_+_183186238 | 1.01 |
ENST00000493293.5
ENST00000264144.5 |
LAMC2
|
laminin subunit gamma 2 |
| chr7_-_108456378 | 0.99 |
ENST00000613830.4
ENST00000413765.6 ENST00000379028.8 |
NRCAM
|
neuronal cell adhesion molecule |
| chr16_+_86578543 | 0.95 |
ENST00000320241.5
|
FOXL1
|
forkhead box L1 |
| chr2_-_132670194 | 0.94 |
ENST00000397463.3
|
LYPD1
|
LY6/PLAUR domain containing 1 |
| chr6_-_29633056 | 0.94 |
ENST00000377016.8
|
GABBR1
|
gamma-aminobutyric acid type B receptor subunit 1 |
| chr6_-_29633171 | 0.90 |
ENST00000377034.9
|
GABBR1
|
gamma-aminobutyric acid type B receptor subunit 1 |
| chr12_-_122500947 | 0.90 |
ENST00000672018.1
|
ZCCHC8
|
zinc finger CCHC-type containing 8 |
| chr1_-_150235972 | 0.89 |
ENST00000534220.1
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chr21_-_26845402 | 0.89 |
ENST00000284984.8
ENST00000676955.1 |
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif 1 |
| chr5_-_151080978 | 0.87 |
ENST00000520931.5
ENST00000521591.6 ENST00000520695.5 ENST00000610535.5 ENST00000518977.5 ENST00000389378.6 ENST00000610874.4 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr1_-_212699817 | 0.86 |
ENST00000243440.2
|
BATF3
|
basic leucine zipper ATF-like transcription factor 3 |
| chrX_+_12791353 | 0.85 |
ENST00000380663.7
ENST00000398491.6 ENST00000380668.10 ENST00000489404.5 |
PRPS2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chr22_-_27801712 | 0.84 |
ENST00000302326.5
|
MN1
|
MN1 proto-oncogene, transcriptional regulator |
| chr16_+_31472130 | 0.82 |
ENST00000394863.8
ENST00000565360.5 ENST00000361773.7 |
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1 |
| chrX_-_109733181 | 0.82 |
ENST00000673016.1
|
ACSL4
|
acyl-CoA synthetase long chain family member 4 |
| chr12_-_122500924 | 0.80 |
ENST00000633063.3
|
ZCCHC8
|
zinc finger CCHC-type containing 8 |
| chr12_-_122500520 | 0.75 |
ENST00000540586.1
ENST00000543897.5 |
ZCCHC8
|
zinc finger CCHC-type containing 8 |
| chr2_-_207165923 | 0.71 |
ENST00000309446.11
|
KLF7
|
Kruppel like factor 7 |
| chr19_-_3546306 | 0.71 |
ENST00000398558.8
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chrX_-_109733249 | 0.71 |
ENST00000469796.7
ENST00000672401.1 ENST00000671846.1 |
ACSL4
|
acyl-CoA synthetase long chain family member 4 |
| chr4_-_148442342 | 0.71 |
ENST00000358102.8
|
NR3C2
|
nuclear receptor subfamily 3 group C member 2 |
| chr6_+_43771960 | 0.63 |
ENST00000230480.10
|
VEGFA
|
vascular endothelial growth factor A |
| chr11_+_111912725 | 0.63 |
ENST00000304298.4
|
HSPB2
|
heat shock protein family B (small) member 2 |
| chr1_-_156500723 | 0.62 |
ENST00000368240.6
|
MEF2D
|
myocyte enhancer factor 2D |
| chrX_-_109733220 | 0.61 |
ENST00000672282.1
ENST00000340800.7 |
ACSL4
|
acyl-CoA synthetase long chain family member 4 |
| chr6_+_162727941 | 0.61 |
ENST00000366888.6
|
PACRG
|
parkin coregulated |
| chr17_-_41140487 | 0.61 |
ENST00000345847.4
|
KRTAP4-6
|
keratin associated protein 4-6 |
| chr12_-_6689359 | 0.60 |
ENST00000683879.1
|
ZNF384
|
zinc finger protein 384 |
| chr15_+_40953463 | 0.59 |
ENST00000617768.5
|
CHAC1
|
ChaC glutathione specific gamma-glutamylcyclotransferase 1 |
| chrX_-_108736556 | 0.59 |
ENST00000372129.4
|
IRS4
|
insulin receptor substrate 4 |
| chr12_-_6689244 | 0.59 |
ENST00000361959.7
ENST00000436774.6 ENST00000544482.1 |
ZNF384
|
zinc finger protein 384 |
| chrX_-_109733292 | 0.59 |
ENST00000682031.1
ENST00000502391.6 ENST00000508092.5 ENST00000348502.10 |
ACSL4
|
acyl-CoA synthetase long chain family member 4 |
| chr12_-_6689450 | 0.58 |
ENST00000355772.8
ENST00000417772.7 ENST00000319770.7 ENST00000396801.7 |
ZNF384
|
zinc finger protein 384 |
| chr8_-_93740718 | 0.55 |
ENST00000519109.6
|
RBM12B
|
RNA binding motif protein 12B |
| chr5_+_157731400 | 0.53 |
ENST00000231198.12
|
THG1L
|
tRNA-histidine guanylyltransferase 1 like |
| chr1_-_41662298 | 0.52 |
ENST00000643665.1
|
HIVEP3
|
HIVEP zinc finger 3 |
| chr5_-_122077152 | 0.52 |
ENST00000513319.5
ENST00000503759.5 |
LOX
|
lysyl oxidase |
| chr20_+_33731976 | 0.52 |
ENST00000375200.6
|
ZNF341
|
zinc finger protein 341 |
| chr7_+_120273129 | 0.48 |
ENST00000331113.9
|
KCND2
|
potassium voltage-gated channel subfamily D member 2 |
| chr15_+_83447411 | 0.47 |
ENST00000324537.5
|
SH3GL3
|
SH3 domain containing GRB2 like 3, endophilin A3 |
| chr11_-_30586272 | 0.47 |
ENST00000448418.6
|
MPPED2
|
metallophosphoesterase domain containing 2 |
| chr10_-_93601228 | 0.47 |
ENST00000371464.8
|
RBP4
|
retinol binding protein 4 |
| chr12_+_4269771 | 0.46 |
ENST00000676411.1
|
CCND2
|
cyclin D2 |
| chr1_-_156500763 | 0.45 |
ENST00000348159.9
ENST00000489057.1 |
MEF2D
|
myocyte enhancer factor 2D |
| chr20_-_31722854 | 0.45 |
ENST00000307677.5
|
BCL2L1
|
BCL2 like 1 |
| chr19_+_1067272 | 0.45 |
ENST00000590214.5
|
ARHGAP45
|
Rho GTPase activating protein 45 |
| chr8_-_70669142 | 0.44 |
ENST00000522447.5
ENST00000276590.5 |
LACTB2
|
lactamase beta 2 |
| chr19_+_1067144 | 0.44 |
ENST00000313093.7
|
ARHGAP45
|
Rho GTPase activating protein 45 |
| chr19_+_1067493 | 0.43 |
ENST00000586866.5
|
ARHGAP45
|
Rho GTPase activating protein 45 |
| chrX_-_108736327 | 0.43 |
ENST00000564206.2
|
IRS4
|
insulin receptor substrate 4 |
| chr22_-_50532077 | 0.43 |
ENST00000428989.3
ENST00000403326.5 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr20_-_4015389 | 0.43 |
ENST00000336095.10
|
RNF24
|
ring finger protein 24 |
| chr2_+_26692686 | 0.42 |
ENST00000620977.1
ENST00000302909.4 |
KCNK3
|
potassium two pore domain channel subfamily K member 3 |
| chr1_-_171652675 | 0.42 |
ENST00000037502.11
|
MYOC
|
myocilin |
| chr5_-_149379286 | 0.42 |
ENST00000261796.4
|
IL17B
|
interleukin 17B |
| chr7_-_113086815 | 0.42 |
ENST00000424100.2
|
GPR85
|
G protein-coupled receptor 85 |
| chr10_+_97584314 | 0.42 |
ENST00000370647.8
|
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr5_-_142325001 | 0.41 |
ENST00000344120.4
ENST00000434127.3 |
SPRY4
|
sprouty RTK signaling antagonist 4 |
| chr20_-_31722949 | 0.41 |
ENST00000376055.9
|
BCL2L1
|
BCL2 like 1 |
| chr2_-_208124514 | 0.41 |
ENST00000264376.5
|
CRYGD
|
crystallin gamma D |
| chr11_-_30586866 | 0.41 |
ENST00000528686.2
|
MPPED2
|
metallophosphoesterase domain containing 2 |
| chr17_-_3916455 | 0.40 |
ENST00000225538.4
|
P2RX1
|
purinergic receptor P2X 1 |
| chr1_-_2633011 | 0.39 |
ENST00000378412.8
|
MMEL1
|
membrane metalloendopeptidase like 1 |
| chr12_-_31324129 | 0.39 |
ENST00000454658.6
|
SINHCAF
|
SIN3-HDAC complex associated factor |
| chr11_-_120120880 | 0.39 |
ENST00000526881.1
|
TRIM29
|
tripartite motif containing 29 |
| chr12_+_71754834 | 0.39 |
ENST00000261263.5
|
RAB21
|
RAB21, member RAS oncogene family |
| chr4_+_165378998 | 0.39 |
ENST00000402744.9
|
CPE
|
carboxypeptidase E |
| chr4_+_76949743 | 0.38 |
ENST00000502584.5
ENST00000264893.11 ENST00000510641.5 |
SEPTIN11
|
septin 11 |
| chr1_+_209768482 | 0.38 |
ENST00000367023.5
|
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr11_-_111913195 | 0.37 |
ENST00000531198.5
ENST00000616970.5 ENST00000527899.6 |
CRYAB
|
crystallin alpha B |
| chr11_-_111913134 | 0.37 |
ENST00000533971.2
ENST00000526180.6 |
CRYAB
|
crystallin alpha B |
| chr1_-_43367956 | 0.36 |
ENST00000372458.8
|
ELOVL1
|
ELOVL fatty acid elongase 1 |
| chr5_+_53480619 | 0.36 |
ENST00000396947.7
ENST00000256759.8 |
FST
|
follistatin |
| chr11_-_19202004 | 0.36 |
ENST00000648719.1
|
CSRP3
|
cysteine and glycine rich protein 3 |
| chr20_-_31723491 | 0.36 |
ENST00000676582.1
ENST00000422920.2 |
BCL2L1
|
BCL2 like 1 |
| chr21_-_34887148 | 0.35 |
ENST00000399240.5
|
RUNX1
|
RUNX family transcription factor 1 |
| chr8_+_42152946 | 0.35 |
ENST00000518421.5
ENST00000174653.3 ENST00000396926.8 ENST00000521280.5 ENST00000522288.5 |
AP3M2
|
adaptor related protein complex 3 subunit mu 2 |
| chr4_+_76949807 | 0.35 |
ENST00000505788.5
ENST00000510515.5 ENST00000504637.5 |
SEPTIN11
|
septin 11 |
| chr3_+_66998297 | 0.35 |
ENST00000484414.5
ENST00000460576.5 ENST00000417314.2 |
KBTBD8
|
kelch repeat and BTB domain containing 8 |
| chr5_-_139404050 | 0.34 |
ENST00000514983.5
ENST00000507779.2 ENST00000451821.6 ENST00000509959.5 ENST00000302091.9 ENST00000450845.7 |
SPATA24
|
spermatogenesis associated 24 |
| chr1_+_100896060 | 0.34 |
ENST00000370112.8
ENST00000357650.9 |
SLC30A7
|
solute carrier family 30 member 7 |
| chr6_+_101393699 | 0.33 |
ENST00000369134.9
ENST00000684068.1 ENST00000683903.1 ENST00000681975.1 |
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chrX_-_153971169 | 0.33 |
ENST00000369984.4
|
HCFC1
|
host cell factor C1 |
| chr1_-_151826085 | 0.32 |
ENST00000356728.11
|
RORC
|
RAR related orphan receptor C |
| chr16_+_30664334 | 0.32 |
ENST00000287468.5
|
FBRS
|
fibrosin |
| chr7_-_92833896 | 0.32 |
ENST00000265734.8
|
CDK6
|
cyclin dependent kinase 6 |
| chr19_-_6424802 | 0.32 |
ENST00000600480.2
|
KHSRP
|
KH-type splicing regulatory protein |
| chr15_+_81299416 | 0.32 |
ENST00000558332.3
|
IL16
|
interleukin 16 |
| chr6_+_45422564 | 0.31 |
ENST00000625924.1
|
RUNX2
|
RUNX family transcription factor 2 |
| chr6_+_45422485 | 0.31 |
ENST00000359524.7
|
RUNX2
|
RUNX family transcription factor 2 |
| chr3_+_170418856 | 0.31 |
ENST00000064724.8
ENST00000486975.1 |
CLDN11
ENSG00000285218.1
|
claudin 11 novel protein |
| chr3_+_4493442 | 0.31 |
ENST00000456211.8
ENST00000443694.5 ENST00000648266.1 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor type 1 |
| chr15_+_40569290 | 0.30 |
ENST00000315616.12
ENST00000559271.1 ENST00000616318.1 |
RPUSD2
|
RNA pseudouridine synthase domain containing 2 |
| chr15_+_60004305 | 0.30 |
ENST00000396057.6
|
FOXB1
|
forkhead box B1 |
| chr11_+_118607598 | 0.30 |
ENST00000600882.6
ENST00000356063.9 |
PHLDB1
|
pleckstrin homology like domain family B member 1 |
| chr3_-_8644782 | 0.30 |
ENST00000544814.6
|
SSUH2
|
ssu-2 homolog |
| chr1_-_43368039 | 0.29 |
ENST00000413844.3
|
ELOVL1
|
ELOVL fatty acid elongase 1 |
| chr2_-_212538766 | 0.29 |
ENST00000342788.9
|
ERBB4
|
erb-b2 receptor tyrosine kinase 4 |
| chr10_-_84241538 | 0.29 |
ENST00000372105.4
|
LRIT1
|
leucine rich repeat, Ig-like and transmembrane domains 1 |
| chr22_-_50532137 | 0.29 |
ENST00000405135.5
ENST00000401779.5 ENST00000682240.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr3_+_4493340 | 0.28 |
ENST00000357086.10
ENST00000354582.12 ENST00000649015.2 ENST00000467056.6 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor type 1 |
| chr17_+_7307579 | 0.28 |
ENST00000572815.5
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr8_-_22156789 | 0.27 |
ENST00000306317.7
|
LGI3
|
leucine rich repeat LGI family member 3 |
| chr9_-_95507416 | 0.27 |
ENST00000429896.6
|
PTCH1
|
patched 1 |
| chr8_-_22156741 | 0.27 |
ENST00000424267.6
|
LGI3
|
leucine rich repeat LGI family member 3 |
| chr10_-_35090237 | 0.26 |
ENST00000673636.1
ENST00000374749.8 ENST00000374748.5 |
CUL2
|
cullin 2 |
| chr2_-_51032151 | 0.26 |
ENST00000628515.2
|
NRXN1
|
neurexin 1 |
| chr15_+_83447328 | 0.26 |
ENST00000427482.7
|
SH3GL3
|
SH3 domain containing GRB2 like 3, endophilin A3 |
| chr17_-_76537699 | 0.26 |
ENST00000293230.10
|
CYGB
|
cytoglobin |
| chr2_+_113890039 | 0.26 |
ENST00000443297.5
ENST00000263238.7 ENST00000415792.5 |
ACTR3
|
actin related protein 3 |
| chr10_-_35090545 | 0.25 |
ENST00000374751.7
ENST00000626172.2 |
CUL2
|
cullin 2 |
| chr9_-_35958154 | 0.25 |
ENST00000341959.2
|
OR2S2
|
olfactory receptor family 2 subfamily S member 2 |
| chr5_-_136365476 | 0.25 |
ENST00000378459.7
ENST00000502753.4 ENST00000513104.6 ENST00000352189.8 |
TRPC7
|
transient receptor potential cation channel subfamily C member 7 |
| chr17_+_7307531 | 0.25 |
ENST00000576930.5
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr3_+_4493471 | 0.25 |
ENST00000544951.6
ENST00000650294.1 |
ITPR1
|
inositol 1,4,5-trisphosphate receptor type 1 |
| chr9_-_127980976 | 0.25 |
ENST00000373095.6
|
FAM102A
|
family with sequence similarity 102 member A |
| chr17_-_42683057 | 0.25 |
ENST00000591765.1
|
CCR10
|
C-C motif chemokine receptor 10 |
| chr9_+_128456135 | 0.24 |
ENST00000604420.5
ENST00000448249.7 ENST00000393527.7 |
ODF2
|
outer dense fiber of sperm tails 2 |
| chr1_-_39639626 | 0.24 |
ENST00000372852.4
|
HEYL
|
hes related family bHLH transcription factor with YRPW motif like |
| chr9_+_128456418 | 0.24 |
ENST00000434106.7
ENST00000546203.5 ENST00000446274.5 ENST00000421776.6 ENST00000432065.6 |
ODF2
|
outer dense fiber of sperm tails 2 |
| chr14_-_24114913 | 0.24 |
ENST00000561028.6
ENST00000558280.1 |
NRL
|
neural retina leucine zipper |
| chr17_-_7234262 | 0.23 |
ENST00000575756.5
ENST00000575458.5 |
DVL2
|
dishevelled segment polarity protein 2 |
| chr2_-_212538841 | 0.23 |
ENST00000436443.5
|
ERBB4
|
erb-b2 receptor tyrosine kinase 4 |
| chr1_-_150235995 | 0.23 |
ENST00000436748.6
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chr15_-_100602157 | 0.23 |
ENST00000559577.5
ENST00000561308.5 ENST00000560133.5 ENST00000560941.5 ENST00000314742.13 ENST00000559736.5 ENST00000560272.1 |
LINS1
|
lines homolog 1 |
| chr11_-_30586344 | 0.23 |
ENST00000358117.10
|
MPPED2
|
metallophosphoesterase domain containing 2 |
| chr19_+_19538248 | 0.22 |
ENST00000586018.5
ENST00000291495.5 |
CILP2
|
cartilage intermediate layer protein 2 |
| chr14_-_106211453 | 0.22 |
ENST00000390606.3
|
IGHV3-20
|
immunoglobulin heavy variable 3-20 |
| chr15_-_78234513 | 0.22 |
ENST00000558130.1
ENST00000258873.9 |
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr9_+_128456899 | 0.22 |
ENST00000372791.7
|
ODF2
|
outer dense fiber of sperm tails 2 |
| chr8_+_30387064 | 0.22 |
ENST00000523115.5
ENST00000519647.5 |
RBPMS
|
RNA binding protein, mRNA processing factor |
| chr11_+_118607579 | 0.22 |
ENST00000530708.4
|
PHLDB1
|
pleckstrin homology like domain family B member 1 |
| chr2_+_178112416 | 0.21 |
ENST00000286070.10
ENST00000616198.4 |
RBM45
|
RNA binding motif protein 45 |
| chr18_-_12884151 | 0.21 |
ENST00000591115.5
ENST00000309660.10 ENST00000353319.8 ENST00000327283.7 |
PTPN2
|
protein tyrosine phosphatase non-receptor type 2 |
| chr1_-_43367689 | 0.21 |
ENST00000621943.4
|
ELOVL1
|
ELOVL fatty acid elongase 1 |
| chr14_-_64504570 | 0.20 |
ENST00000394715.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chrX_-_23743201 | 0.20 |
ENST00000492081.1
ENST00000379303.10 ENST00000336430.11 |
ACOT9
|
acyl-CoA thioesterase 9 |
| chrX_+_71223216 | 0.20 |
ENST00000361726.7
|
GJB1
|
gap junction protein beta 1 |
| chr11_+_76782250 | 0.20 |
ENST00000533752.1
ENST00000612930.1 |
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr3_+_50269140 | 0.20 |
ENST00000616701.5
ENST00000433753.4 ENST00000611067.4 |
SEMA3B
|
semaphorin 3B |
| chr19_+_4969105 | 0.20 |
ENST00000611640.4
ENST00000159111.9 ENST00000588337.5 ENST00000381759.8 |
KDM4B
|
lysine demethylase 4B |
| chr16_+_33827140 | 0.20 |
ENST00000562905.2
|
IGHV3OR16-13
|
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
| chr9_+_128411715 | 0.20 |
ENST00000420034.5
ENST00000372842.5 |
CERCAM
|
cerebral endothelial cell adhesion molecule |
| chr20_-_52105644 | 0.19 |
ENST00000371523.8
|
ZFP64
|
ZFP64 zinc finger protein |
| chr10_+_24466487 | 0.19 |
ENST00000396446.5
ENST00000396445.5 ENST00000376451.4 |
KIAA1217
|
KIAA1217 |
| chr19_-_35134861 | 0.19 |
ENST00000591633.2
|
LGI4
|
leucine rich repeat LGI family member 4 |
| chr1_-_18859682 | 0.19 |
ENST00000375371.3
|
TAS1R2
|
taste 1 receptor member 2 |
| chr1_-_150235943 | 0.19 |
ENST00000533654.5
|
ANP32E
|
acidic nuclear phosphoprotein 32 family member E |
| chr22_-_50532489 | 0.19 |
ENST00000329363.9
ENST00000437588.2 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr19_-_6424772 | 0.18 |
ENST00000619396.4
ENST00000398148.7 |
KHSRP
|
KH-type splicing regulatory protein |
| chr7_+_149715003 | 0.18 |
ENST00000496259.5
ENST00000319551.12 |
KRBA1
|
KRAB-A domain containing 1 |
| chr3_+_121567924 | 0.18 |
ENST00000334384.5
|
ARGFX
|
arginine-fifty homeobox |
| chr7_-_92245795 | 0.18 |
ENST00000412043.6
ENST00000430102.5 ENST00000425073.1 ENST00000394505.7 ENST00000394503.6 ENST00000454017.5 ENST00000440209.5 ENST00000413688.5 ENST00000452773.5 ENST00000433016.5 ENST00000422347.5 ENST00000458493.5 ENST00000425919.5 ENST00000650585.1 |
KRIT1
ENSG00000285953.1
|
KRIT1 ankyrin repeat containing novel protein |
| chr3_+_10026409 | 0.18 |
ENST00000287647.7
ENST00000676013.1 ENST00000675286.1 ENST00000419585.5 |
FANCD2
|
FA complementation group D2 |
| chr14_-_88323238 | 0.18 |
ENST00000319231.10
|
KCNK10
|
potassium two pore domain channel subfamily K member 10 |
| chr1_+_154993581 | 0.17 |
ENST00000392487.1
|
LENEP
|
lens epithelial protein |
| chr5_+_161848314 | 0.17 |
ENST00000437025.6
|
GABRA1
|
gamma-aminobutyric acid type A receptor subunit alpha1 |
| chr4_-_175812746 | 0.17 |
ENST00000393658.6
|
GPM6A
|
glycoprotein M6A |
| chr5_+_163437569 | 0.17 |
ENST00000512163.5
ENST00000393929.5 ENST00000510097.5 ENST00000340828.7 ENST00000511490.4 ENST00000510664.5 |
CCNG1
|
cyclin G1 |
| chr19_+_42268505 | 0.17 |
ENST00000572681.6
|
CIC
|
capicua transcriptional repressor |
| chr7_+_90403386 | 0.17 |
ENST00000287916.8
ENST00000394604.5 ENST00000496677.6 ENST00000394605.2 ENST00000480135.1 |
CLDN12
ENSG00000273299.1
|
claudin 12 novel transcript |
| chr11_-_111911759 | 0.17 |
ENST00000650687.2
|
CRYAB
|
crystallin alpha B |
| chr7_-_5423826 | 0.17 |
ENST00000430969.6
|
TNRC18
|
trinucleotide repeat containing 18 |
| chr9_+_128456006 | 0.17 |
ENST00000351030.7
ENST00000372814.7 |
ODF2
|
outer dense fiber of sperm tails 2 |
| chr9_-_95317671 | 0.16 |
ENST00000490972.7
ENST00000647778.1 ENST00000649611.1 ENST00000289081.8 |
FANCC
|
FA complementation group C |
| chr9_+_113349514 | 0.16 |
ENST00000374183.5
|
BSPRY
|
B-box and SPRY domain containing |
| chr7_+_150405146 | 0.16 |
ENST00000498682.3
ENST00000641717.1 |
ENSG00000284691.1
|
novel zinc finger protein |
| chr1_-_156490599 | 0.16 |
ENST00000360595.7
|
MEF2D
|
myocyte enhancer factor 2D |
| chr9_+_33264848 | 0.16 |
ENST00000419016.6
|
CHMP5
|
charged multivesicular body protein 5 |
| chr16_+_53130921 | 0.16 |
ENST00000564845.5
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr11_-_75668566 | 0.16 |
ENST00000526740.3
|
MAP6
|
microtubule associated protein 6 |
| chr10_+_102644990 | 0.16 |
ENST00000645961.1
|
TRIM8
|
tripartite motif containing 8 |
| chr16_+_30023198 | 0.16 |
ENST00000681219.1
ENST00000300575.6 |
C16orf92
|
chromosome 16 open reading frame 92 |
| chr1_+_150549384 | 0.16 |
ENST00000369041.9
ENST00000271643.9 |
ADAMTSL4
|
ADAMTS like 4 |
| chr4_+_71187269 | 0.15 |
ENST00000425175.5
ENST00000351898.10 |
SLC4A4
|
solute carrier family 4 member 4 |
| chr19_-_12957198 | 0.15 |
ENST00000316939.3
|
GADD45GIP1
|
GADD45G interacting protein 1 |
| chr9_-_127877665 | 0.15 |
ENST00000644144.2
|
AK1
|
adenylate kinase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.7 | 5.3 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.4 | 1.5 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.3 | 4.4 | GO:0038129 | ERBB3 signaling pathway(GO:0038129) |
| 0.3 | 1.5 | GO:0072144 | glomerular mesangial cell development(GO:0072144) mesenchyme migration(GO:0090131) |
| 0.3 | 0.9 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.2 | 3.7 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 | 2.7 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.2 | 4.1 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.2 | 0.6 | GO:1903572 | coronary vein morphogenesis(GO:0003169) regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.2 | 3.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 5.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 1.0 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.2 | 0.5 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.2 | 0.9 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.2 | 1.4 | GO:1904628 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.2 | 1.2 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.1 | 1.6 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 0.3 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.1 | 0.8 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 0.8 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 3.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.5 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.1 | 1.2 | GO:0007512 | adult heart development(GO:0007512) |
| 0.1 | 0.4 | GO:0002442 | serotonin production involved in inflammatory response(GO:0002351) serotonin secretion involved in inflammatory response(GO:0002442) serotonin secretion by platelet(GO:0002554) |
| 0.1 | 0.9 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.4 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.8 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.4 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.1 | 0.6 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.2 | GO:1902232 | regulation of interleukin-4-mediated signaling pathway(GO:1902214) negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) regulation of positive thymic T cell selection(GO:1902232) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.1 | 0.5 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.2 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 1.7 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 0.3 | GO:0061030 | axon target recognition(GO:0007412) epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.1 | 0.1 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.0 | 0.2 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.2 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 2.5 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.3 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.5 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.3 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.5 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.2 | GO:1904327 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.3 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.2 | GO:0072014 | proximal tubule development(GO:0072014) |
| 0.0 | 1.0 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.2 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.2 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.5 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.6 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.1 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.0 | 0.1 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.0 | 0.9 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 1.7 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.3 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.3 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.0 | 0.4 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.3 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.4 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.4 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.3 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0002584 | negative regulation of antigen processing and presentation of peptide antigen(GO:0002584) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.0 | 0.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.2 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.2 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.1 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.0 | 0.3 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0019086 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) |
| 0.0 | 0.5 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.3 | GO:0036315 | cellular response to sterol(GO:0036315) |
| 0.0 | 0.3 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 1.2 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 0.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.1 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 0.2 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.0 | 0.1 | GO:1903874 | ferrous iron transport(GO:0015684) ferrous iron transmembrane transport(GO:1903874) |
| 0.0 | 0.3 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.3 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.1 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.4 | 3.0 | GO:0042825 | TAP complex(GO:0042825) |
| 0.3 | 1.5 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.2 | 3.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 0.9 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 2.7 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 5.3 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.7 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 1.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 1.0 | GO:0043256 | laminin complex(GO:0043256) |
| 0.1 | 1.6 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.5 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 3.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 2.0 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.5 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 0.9 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.4 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.8 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.4 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.5 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 2.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.5 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 2.2 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.2 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.7 | 3.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.4 | 3.0 | GO:0023029 | peptide-transporting ATPase activity(GO:0015440) MHC class Ib protein binding(GO:0023029) TAP2 binding(GO:0046979) |
| 0.4 | 1.4 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.3 | 1.6 | GO:0005119 | smoothened binding(GO:0005119) hedgehog family protein binding(GO:0097108) |
| 0.3 | 0.9 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.3 | 5.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.3 | 5.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 4.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.6 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.9 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.5 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.1 | 0.6 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 3.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 2.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.8 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 1.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.9 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.3 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.1 | 0.5 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 0.2 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.4 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.4 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.5 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.3 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.4 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.3 | GO:0046972 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 1.0 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 1.0 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.5 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 3.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.5 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 1.5 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.9 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.2 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.3 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.7 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.7 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.8 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 2.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 3.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 3.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 5.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 4.5 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 3.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 3.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.7 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 1.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 2.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 1.2 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.6 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.0 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |