Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
Results for ZKSCAN3
Z-value: 0.35
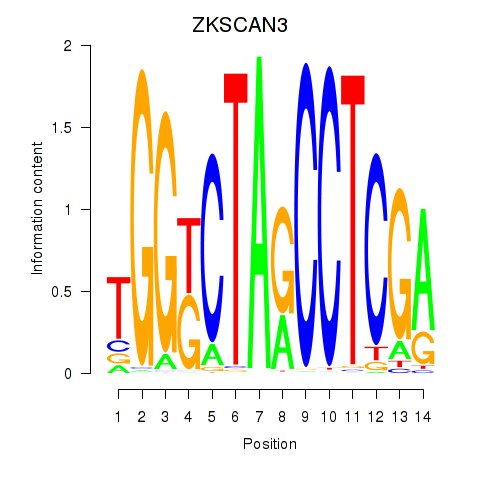
Transcription factors associated with ZKSCAN3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZKSCAN3
|
ENSG00000189298.14 | ZKSCAN3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZKSCAN3 | hg38_v1_chr6_+_28349907_28349983 | -0.09 | 6.9e-01 | Click! |
Activity profile of ZKSCAN3 motif
Sorted Z-values of ZKSCAN3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZKSCAN3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_1044542 | 0.98 |
ENST00000444847.2
|
GPR146
|
G protein-coupled receptor 146 |
| chr19_+_39296399 | 0.82 |
ENST00000333625.3
|
IFNL1
|
interferon lambda 1 |
| chr2_+_190880809 | 0.80 |
ENST00000320717.8
|
GLS
|
glutaminase |
| chr7_+_155298561 | 0.77 |
ENST00000476756.1
|
INSIG1
|
insulin induced gene 1 |
| chr17_+_34270213 | 0.75 |
ENST00000378569.2
ENST00000394627.5 ENST00000394630.3 |
CCL7
|
C-C motif chemokine ligand 7 |
| chr19_-_39245006 | 0.51 |
ENST00000413851.3
ENST00000613087.4 |
IFNL3
|
interferon lambda 3 |
| chr1_+_65792889 | 0.50 |
ENST00000341517.9
|
PDE4B
|
phosphodiesterase 4B |
| chr3_-_122793772 | 0.47 |
ENST00000306103.3
|
HSPBAP1
|
HSPB1 associated protein 1 |
| chr6_+_158560057 | 0.42 |
ENST00000684151.1
|
TMEM181
|
transmembrane protein 181 |
| chr12_+_109098118 | 0.41 |
ENST00000336865.6
|
UNG
|
uracil DNA glycosylase |
| chr1_+_93448108 | 0.41 |
ENST00000271234.13
ENST00000260506.12 |
FNBP1L
|
formin binding protein 1 like |
| chr1_+_3682306 | 0.40 |
ENST00000603362.5
ENST00000604479.5 |
TP73
|
tumor protein p73 |
| chr1_+_93448155 | 0.33 |
ENST00000370253.6
|
FNBP1L
|
formin binding protein 1 like |
| chrX_+_151694967 | 0.32 |
ENST00000448726.5
ENST00000538575.5 |
PRRG3
|
proline rich and Gla domain 3 |
| chr16_-_28495519 | 0.31 |
ENST00000569430.7
|
CLN3
|
CLN3 lysosomal/endosomal transmembrane protein, battenin |
| chr10_+_75431605 | 0.30 |
ENST00000611255.5
|
LRMDA
|
leucine rich melanocyte differentiation associated |
| chr5_+_176810552 | 0.25 |
ENST00000329542.9
|
UNC5A
|
unc-5 netrin receptor A |
| chr21_+_46001300 | 0.24 |
ENST00000612273.2
ENST00000682634.1 |
COL6A1
|
collagen type VI alpha 1 chain |
| chr5_+_176810498 | 0.24 |
ENST00000509580.2
|
UNC5A
|
unc-5 netrin receptor A |
| chr6_+_42929430 | 0.18 |
ENST00000372836.5
|
CNPY3
|
canopy FGF signaling regulator 3 |
| chr6_+_32968557 | 0.16 |
ENST00000374825.9
|
BRD2
|
bromodomain containing 2 |
| chr17_-_18644418 | 0.15 |
ENST00000575220.5
ENST00000405044.6 ENST00000573652.1 |
TBC1D28
|
TBC1 domain family member 28 |
| chr2_+_190880834 | 0.13 |
ENST00000338435.8
|
GLS
|
glutaminase |
| chr2_-_166375901 | 0.12 |
ENST00000303354.11
ENST00000645907.1 ENST00000642356.2 ENST00000452182.2 |
SCN9A
|
sodium voltage-gated channel alpha subunit 9 |
| chr11_-_119346695 | 0.11 |
ENST00000619721.6
|
MFRP
|
membrane frizzled-related protein |
| chrX_+_86148441 | 0.10 |
ENST00000373125.9
ENST00000373131.5 |
DACH2
|
dachshund family transcription factor 2 |
| chr2_-_166375969 | 0.09 |
ENST00000454569.6
ENST00000409672.5 |
SCN9A
|
sodium voltage-gated channel alpha subunit 9 |
| chr1_+_226062704 | 0.09 |
ENST00000366814.3
ENST00000366815.10 ENST00000655399.1 ENST00000667897.1 |
H3-3A
|
H3.3 histone A |
| chr7_-_151210488 | 0.08 |
ENST00000644661.2
|
H2BE1
|
H2B.E variant histone 1 |
| chr1_-_19484635 | 0.07 |
ENST00000433834.5
|
CAPZB
|
capping actin protein of muscle Z-line subunit beta |
| chr1_-_86156737 | 0.07 |
ENST00000370571.7
|
COL24A1
|
collagen type XXIV alpha 1 chain |
| chr14_-_106389858 | 0.06 |
ENST00000390617.2
|
IGHV3-35
|
immunoglobulin heavy variable 3-35 (non-functional) |
| chr20_-_59042748 | 0.06 |
ENST00000355937.9
ENST00000371033.9 |
PRELID3B
|
PRELI domain containing 3B |
| chr22_+_49918626 | 0.05 |
ENST00000328268.9
ENST00000404488.7 |
CRELD2
|
cysteine rich with EGF like domains 2 |
| chr22_-_49918404 | 0.05 |
ENST00000330817.11
|
ALG12
|
ALG12 alpha-1,6-mannosyltransferase |
| chr15_-_41332487 | 0.03 |
ENST00000560640.1
ENST00000220514.8 |
OIP5
|
Opa interacting protein 5 |
| chr22_+_49918733 | 0.03 |
ENST00000407217.7
ENST00000403427.3 |
CRELD2
|
cysteine rich with EGF like domains 2 |
| chr6_-_31540536 | 0.02 |
ENST00000376177.6
|
DDX39B
|
DExD-box helicase 39B |
| chr19_-_4182527 | 0.01 |
ENST00000601571.1
ENST00000601488.5 ENST00000305232.10 ENST00000337491.7 |
SIRT6
|
sirtuin 6 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.2 | 0.9 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.8 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 0.4 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 0.8 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.1 | 0.3 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 0.5 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.7 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.5 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.4 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.4 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.2 | 0.8 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 0.8 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.4 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |