Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
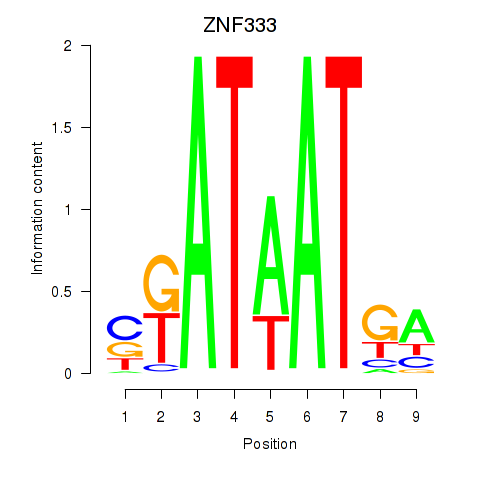
Results for ZNF333
Z-value: 0.61
Transcription factors associated with ZNF333
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF333
|
ENSG00000160961.12 | ZNF333 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF333 | hg38_v1_chr19_+_14690037_14690105 | 0.52 | 7.5e-03 | Click! |
Activity profile of ZNF333 motif
Sorted Z-values of ZNF333 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF333
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_116030735 | 1.53 |
ENST00000393485.5
|
TFEC
|
transcription factor EC |
| chr7_-_116030750 | 1.39 |
ENST00000265440.12
ENST00000320239.11 |
TFEC
|
transcription factor EC |
| chr16_+_84294853 | 0.73 |
ENST00000219454.10
|
WFDC1
|
WAP four-disulfide core domain 1 |
| chr16_+_84294823 | 0.71 |
ENST00000568638.1
|
WFDC1
|
WAP four-disulfide core domain 1 |
| chr6_+_29306626 | 0.71 |
ENST00000377160.4
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1 |
| chr22_-_30246739 | 0.64 |
ENST00000403987.3
ENST00000249075.4 |
LIF
|
LIF interleukin 6 family cytokine |
| chr3_-_112845950 | 0.63 |
ENST00000398214.5
|
CD200R1L
|
CD200 receptor 1 like |
| chr3_-_116444983 | 0.56 |
ENST00000333617.8
|
LSAMP
|
limbic system associated membrane protein |
| chr12_-_48999363 | 0.51 |
ENST00000421952.3
|
DDN
|
dendrin |
| chr6_+_29301701 | 0.51 |
ENST00000641895.1
|
OR14J1
|
olfactory receptor family 14 subfamily J member 1 |
| chr3_+_173398438 | 0.49 |
ENST00000457714.5
|
NLGN1
|
neuroligin 1 |
| chr20_+_59940362 | 0.49 |
ENST00000360816.8
|
FAM217B
|
family with sequence similarity 217 member B |
| chr12_+_112125531 | 0.48 |
ENST00000549358.5
ENST00000257604.9 ENST00000548092.5 ENST00000412615.7 ENST00000552896.1 |
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr8_-_133102477 | 0.43 |
ENST00000522119.5
ENST00000523610.5 ENST00000338087.10 ENST00000521302.5 ENST00000519558.5 ENST00000519747.5 ENST00000517648.5 |
SLA
|
Src like adaptor |
| chr6_+_132570322 | 0.42 |
ENST00000275198.1
|
TAAR6
|
trace amine associated receptor 6 |
| chr11_-_72793636 | 0.41 |
ENST00000538536.5
ENST00000543304.5 ENST00000540587.1 |
STARD10
|
StAR related lipid transfer domain containing 10 |
| chr3_+_35679614 | 0.40 |
ENST00000474696.5
ENST00000412048.5 ENST00000396482.6 ENST00000432682.5 |
ARPP21
|
cAMP regulated phosphoprotein 21 |
| chr3_-_112975018 | 0.39 |
ENST00000471858.5
ENST00000308611.8 ENST00000295863.4 |
CD200R1
|
CD200 receptor 1 |
| chr11_+_5449323 | 0.37 |
ENST00000641930.1
|
OR51I2
|
olfactory receptor family 51 subfamily I member 2 |
| chr2_+_232662733 | 0.36 |
ENST00000410095.5
ENST00000611312.1 |
EFHD1
|
EF-hand domain family member D1 |
| chr3_+_35679690 | 0.35 |
ENST00000413378.5
ENST00000417925.5 |
ARPP21
|
cAMP regulated phosphoprotein 21 |
| chr20_-_59940289 | 0.35 |
ENST00000370996.5
|
PPP1R3D
|
protein phosphatase 1 regulatory subunit 3D |
| chrX_-_21758097 | 0.33 |
ENST00000379494.4
|
SMPX
|
small muscle protein X-linked |
| chr7_-_78771265 | 0.33 |
ENST00000630991.2
ENST00000629359.2 |
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr2_+_102311502 | 0.31 |
ENST00000404917.6
ENST00000410040.5 |
IL1RL1
IL18R1
|
interleukin 1 receptor like 1 interleukin 18 receptor 1 |
| chr4_-_46909206 | 0.30 |
ENST00000396533.5
|
COX7B2
|
cytochrome c oxidase subunit 7B2 |
| chr1_+_19312341 | 0.29 |
ENST00000400548.6
|
SLC66A1
|
solute carrier family 66 member 1 |
| chr1_+_19312296 | 0.29 |
ENST00000375155.7
ENST00000375153.8 |
SLC66A1
|
solute carrier family 66 member 1 |
| chr10_+_52314272 | 0.28 |
ENST00000373970.4
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr6_+_131135408 | 0.28 |
ENST00000431975.7
ENST00000683794.1 |
AKAP7
|
A-kinase anchoring protein 7 |
| chr5_+_173918186 | 0.28 |
ENST00000657000.1
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr1_+_196888014 | 0.27 |
ENST00000367416.6
ENST00000608469.6 ENST00000251424.8 ENST00000367418.2 |
CFHR4
|
complement factor H related 4 |
| chr15_+_76336755 | 0.26 |
ENST00000290759.9
|
ISL2
|
ISL LIM homeobox 2 |
| chr11_-_72793592 | 0.26 |
ENST00000536377.5
ENST00000359373.9 |
STARD10
ARAP1
|
StAR related lipid transfer domain containing 10 ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr21_-_28992947 | 0.26 |
ENST00000389194.7
ENST00000389195.7 ENST00000614971.4 |
LTN1
|
listerin E3 ubiquitin protein ligase 1 |
| chr2_+_200305976 | 0.26 |
ENST00000358677.9
|
SPATS2L
|
spermatogenesis associated serine rich 2 like |
| chr3_+_98166696 | 0.25 |
ENST00000641450.1
|
OR5H15
|
olfactory receptor family 5 subfamily H member 15 |
| chr5_-_16508858 | 0.24 |
ENST00000684456.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr5_-_16508951 | 0.23 |
ENST00000682628.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr14_+_20110739 | 0.23 |
ENST00000641386.2
ENST00000641633.2 |
OR4K17
|
olfactory receptor family 4 subfamily K member 17 |
| chr7_-_78771108 | 0.23 |
ENST00000626691.2
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr1_-_51297990 | 0.22 |
ENST00000530004.5
|
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr12_-_86838867 | 0.22 |
ENST00000621808.4
|
MGAT4C
|
MGAT4 family member C |
| chr5_-_16508812 | 0.22 |
ENST00000683414.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr1_-_23014024 | 0.22 |
ENST00000440767.2
ENST00000622840.1 |
TEX46
|
testis expressed 46 |
| chr3_+_98130721 | 0.22 |
ENST00000641874.1
|
OR5H1
|
olfactory receptor family 5 subfamily H member 1 |
| chr7_+_141764097 | 0.21 |
ENST00000247879.2
|
TAS2R3
|
taste 2 receptor member 3 |
| chr12_-_52601507 | 0.21 |
ENST00000354310.4
|
KRT72
|
keratin 72 |
| chr12_-_52601458 | 0.20 |
ENST00000537672.6
ENST00000293745.7 |
KRT72
|
keratin 72 |
| chr3_+_14675128 | 0.20 |
ENST00000435614.5
ENST00000253697.8 ENST00000412910.1 |
C3orf20
|
chromosome 3 open reading frame 20 |
| chr5_-_27038576 | 0.19 |
ENST00000511822.1
ENST00000231021.9 |
CDH9
|
cadherin 9 |
| chr1_+_159439722 | 0.19 |
ENST00000641630.1
ENST00000423932.6 |
OR10J1
|
olfactory receptor family 10 subfamily J member 1 |
| chr5_-_139383284 | 0.19 |
ENST00000353963.7
ENST00000348729.8 |
SLC23A1
|
solute carrier family 23 member 1 |
| chrX_-_19970298 | 0.19 |
ENST00000379687.7
ENST00000379682.8 |
BCLAF3
|
BCLAF1 and THRAP3 family member 3 |
| chr5_+_41904329 | 0.19 |
ENST00000381647.7
ENST00000612065.1 |
C5orf51
|
chromosome 5 open reading frame 51 |
| chr1_+_74235377 | 0.18 |
ENST00000326637.8
|
TNNI3K
|
TNNI3 interacting kinase |
| chr11_+_33039996 | 0.18 |
ENST00000432887.5
ENST00000528898.1 ENST00000531632.6 |
TCP11L1
|
t-complex 11 like 1 |
| chr9_-_111036207 | 0.18 |
ENST00000541779.5
|
LPAR1
|
lysophosphatidic acid receptor 1 |
| chr18_+_58341038 | 0.18 |
ENST00000679791.1
|
NEDD4L
|
NEDD4 like E3 ubiquitin protein ligase |
| chr10_+_49299159 | 0.18 |
ENST00000374144.8
|
C10orf71
|
chromosome 10 open reading frame 71 |
| chr2_+_190137760 | 0.18 |
ENST00000396974.2
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr8_+_18391276 | 0.17 |
ENST00000286479.4
ENST00000520116.1 |
NAT2
|
N-acetyltransferase 2 |
| chr4_-_46909235 | 0.16 |
ENST00000505102.1
ENST00000355591.8 |
COX7B2
|
cytochrome c oxidase subunit 7B2 |
| chr13_-_46142834 | 0.16 |
ENST00000674665.1
|
LCP1
|
lymphocyte cytosolic protein 1 |
| chr7_+_90383672 | 0.16 |
ENST00000416322.5
|
CLDN12
|
claudin 12 |
| chr17_+_34961533 | 0.16 |
ENST00000361952.5
|
ZNF830
|
zinc finger protein 830 |
| chr7_+_144069811 | 0.15 |
ENST00000641663.1
|
OR2A25
|
olfactory receptor family 2 subfamily A member 25 |
| chr7_-_5967204 | 0.15 |
ENST00000337579.4
|
RSPH10B
|
radial spoke head 10 homolog B |
| chr7_-_25228485 | 0.14 |
ENST00000222674.2
|
NPVF
|
neuropeptide VF precursor |
| chr10_-_126521439 | 0.14 |
ENST00000284694.11
ENST00000432642.5 |
C10orf90
|
chromosome 10 open reading frame 90 |
| chr7_-_83649097 | 0.14 |
ENST00000643230.2
|
SEMA3E
|
semaphorin 3E |
| chr19_+_926001 | 0.14 |
ENST00000263620.8
|
ARID3A
|
AT-rich interaction domain 3A |
| chr5_-_16508788 | 0.14 |
ENST00000682142.1
|
RETREG1
|
reticulophagy regulator 1 |
| chr10_-_67665642 | 0.14 |
ENST00000682945.1
ENST00000330298.6 ENST00000682758.1 |
CTNNA3
|
catenin alpha 3 |
| chr1_-_36323593 | 0.14 |
ENST00000490466.2
|
EVA1B
|
eva-1 homolog B |
| chr15_-_42491105 | 0.13 |
ENST00000565380.5
ENST00000564754.7 |
ZNF106
|
zinc finger protein 106 |
| chr11_+_18412292 | 0.13 |
ENST00000541669.6
ENST00000280704.8 |
LDHC
|
lactate dehydrogenase C |
| chr5_+_173918216 | 0.13 |
ENST00000519467.1
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr10_-_123891742 | 0.13 |
ENST00000241305.4
ENST00000615851.4 |
CPXM2
|
carboxypeptidase X, M14 family member 2 |
| chr3_+_48241046 | 0.13 |
ENST00000427617.6
ENST00000412564.5 ENST00000354698.8 ENST00000440261.6 |
ZNF589
|
zinc finger protein 589 |
| chr14_+_23372809 | 0.12 |
ENST00000397242.2
ENST00000329715.2 |
IL25
|
interleukin 25 |
| chr8_+_106447918 | 0.12 |
ENST00000442977.6
|
OXR1
|
oxidation resistance 1 |
| chr12_-_6700788 | 0.12 |
ENST00000320591.9
ENST00000534837.6 |
PIANP
|
PILR alpha associated neural protein |
| chrX_-_70260199 | 0.12 |
ENST00000374519.4
|
P2RY4
|
pyrimidinergic receptor P2Y4 |
| chr5_-_16508990 | 0.12 |
ENST00000399793.6
|
RETREG1
|
reticulophagy regulator 1 |
| chr19_-_8896090 | 0.12 |
ENST00000599436.1
|
MUC16
|
mucin 16, cell surface associated |
| chr8_-_30912998 | 0.12 |
ENST00000643185.2
|
TEX15
|
testis expressed 15, meiosis and synapsis associated |
| chr2_+_189674662 | 0.12 |
ENST00000684021.1
|
ANKAR
|
ankyrin and armadillo repeat containing |
| chr17_-_3127551 | 0.11 |
ENST00000328890.3
|
OR1G1
|
olfactory receptor family 1 subfamily G member 1 |
| chr3_+_196867856 | 0.11 |
ENST00000445299.6
ENST00000323460.10 ENST00000419026.5 |
SENP5
|
SUMO specific peptidase 5 |
| chr1_+_218285283 | 0.11 |
ENST00000366932.4
|
RRP15
|
ribosomal RNA processing 15 homolog |
| chr2_-_127220293 | 0.11 |
ENST00000664447.2
ENST00000409327.2 |
CYP27C1
|
cytochrome P450 family 27 subfamily C member 1 |
| chr2_-_2331336 | 0.11 |
ENST00000648933.1
ENST00000644820.1 |
MYT1L
|
myelin transcription factor 1 like |
| chr22_+_22720615 | 0.11 |
ENST00000390309.2
|
IGLV3-19
|
immunoglobulin lambda variable 3-19 |
| chr9_-_19786928 | 0.10 |
ENST00000341998.6
ENST00000286344.3 |
SLC24A2
|
solute carrier family 24 member 2 |
| chr12_-_42484298 | 0.10 |
ENST00000640055.1
ENST00000639566.1 ENST00000455697.6 ENST00000639589.1 |
PRICKLE1
|
prickle planar cell polarity protein 1 |
| chr2_+_200306048 | 0.10 |
ENST00000409988.7
ENST00000409385.5 |
SPATS2L
|
spermatogenesis associated serine rich 2 like |
| chr2_+_88885397 | 0.09 |
ENST00000390243.2
|
IGKV4-1
|
immunoglobulin kappa variable 4-1 |
| chr20_+_13008919 | 0.09 |
ENST00000399002.7
ENST00000434210.5 |
SPTLC3
|
serine palmitoyltransferase long chain base subunit 3 |
| chr7_+_30284574 | 0.09 |
ENST00000323037.5
|
ZNRF2
|
zinc and ring finger 2 |
| chr20_+_1118590 | 0.09 |
ENST00000246015.8
ENST00000335877.11 |
PSMF1
|
proteasome inhibitor subunit 1 |
| chr2_-_213151590 | 0.09 |
ENST00000374319.8
ENST00000457361.5 ENST00000451136.6 ENST00000434687.6 |
IKZF2
|
IKAROS family zinc finger 2 |
| chr6_+_29585121 | 0.09 |
ENST00000641840.1
|
OR2H2
|
olfactory receptor family 2 subfamily H member 2 |
| chrX_+_11293411 | 0.09 |
ENST00000348912.4
ENST00000380714.7 ENST00000380712.7 |
AMELX
|
amelogenin X-linked |
| chr18_+_31376777 | 0.09 |
ENST00000308128.9
ENST00000359747.4 |
DSG4
|
desmoglein 4 |
| chr17_-_62806632 | 0.09 |
ENST00000583803.1
ENST00000456609.6 |
MARCHF10
|
membrane associated ring-CH-type finger 10 |
| chr3_-_42875871 | 0.08 |
ENST00000316161.6
ENST00000437102.1 |
CYP8B1
|
cytochrome P450 family 8 subfamily B member 1 |
| chr17_+_4710622 | 0.08 |
ENST00000574954.5
ENST00000269260.7 ENST00000346341.6 ENST00000572457.5 ENST00000381488.10 ENST00000412477.7 ENST00000571428.5 ENST00000575877.5 |
ARRB2
|
arrestin beta 2 |
| chr14_+_85530163 | 0.08 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr10_-_121596117 | 0.07 |
ENST00000351936.11
|
FGFR2
|
fibroblast growth factor receptor 2 |
| chrY_+_18546691 | 0.07 |
ENST00000309834.8
ENST00000307393.3 ENST00000382856.2 |
HSFY1
|
heat shock transcription factor Y-linked 1 |
| chr3_-_37174578 | 0.07 |
ENST00000336686.9
|
LRRFIP2
|
LRR binding FLII interacting protein 2 |
| chr16_+_53099100 | 0.07 |
ENST00000565832.5
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr15_+_38454103 | 0.07 |
ENST00000397609.6
ENST00000491535.5 |
FAM98B
|
family with sequence similarity 98 member B |
| chr8_-_11868043 | 0.07 |
ENST00000676843.1
ENST00000534510.6 ENST00000676825.1 ENST00000678145.1 ENST00000533455.6 ENST00000353047.11 ENST00000677650.1 ENST00000526195.6 ENST00000676691.1 ENST00000678598.1 ENST00000505496.7 ENST00000527215.7 ENST00000345125.8 ENST00000532656.7 ENST00000678067.1 ENST00000453527.7 ENST00000677415.1 ENST00000530640.7 ENST00000677418.1 ENST00000531089.6 ENST00000677544.1 ENST00000676502.1 ENST00000524500.6 ENST00000677873.1 ENST00000678629.1 ENST00000678929.1 ENST00000677819.1 ENST00000678357.1 ENST00000679051.1 ENST00000677082.1 ENST00000531502.6 ENST00000530296.6 ENST00000534636.6 ENST00000534149.6 ENST00000677366.1 ENST00000676755.1 ENST00000679140.1 ENST00000527243.6 ENST00000677047.1 ENST00000678242.1 |
CTSB
|
cathepsin B |
| chr4_-_26490453 | 0.06 |
ENST00000295589.4
|
CCKAR
|
cholecystokinin A receptor |
| chr2_+_200305873 | 0.06 |
ENST00000439084.5
ENST00000409718.5 |
SPATS2L
|
spermatogenesis associated serine rich 2 like |
| chr14_+_85530127 | 0.06 |
ENST00000330753.6
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr5_-_78985249 | 0.06 |
ENST00000565165.2
|
ARSB
|
arylsulfatase B |
| chr6_-_32763522 | 0.06 |
ENST00000435145.6
ENST00000437316.7 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr2_+_69893940 | 0.06 |
ENST00000244227.8
ENST00000409116.5 |
SNRNP27
|
small nuclear ribonucleoprotein U4/U6.U5 subunit 27 |
| chr16_+_81739027 | 0.06 |
ENST00000566191.5
ENST00000565272.5 ENST00000563954.5 ENST00000565054.5 |
ENSG00000261218.5
PLCG2
|
novel transcript phospholipase C gamma 2 |
| chr19_-_43883964 | 0.05 |
ENST00000587539.2
|
ZNF404
|
zinc finger protein 404 |
| chr2_-_32011001 | 0.05 |
ENST00000404530.6
|
MEMO1
|
mediator of cell motility 1 |
| chr5_+_140827950 | 0.05 |
ENST00000378126.4
ENST00000529310.6 ENST00000527624.1 |
PCDHA6
|
protocadherin alpha 6 |
| chr2_+_200440649 | 0.05 |
ENST00000366118.2
|
SPATS2L
|
spermatogenesis associated serine rich 2 like |
| chr6_-_32941018 | 0.05 |
ENST00000418107.3
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr10_+_116590956 | 0.05 |
ENST00000358834.9
ENST00000528052.5 |
PNLIPRP1
|
pancreatic lipase related protein 1 |
| chrX_-_49264668 | 0.05 |
ENST00000455775.7
ENST00000652559.1 ENST00000376207.10 ENST00000557224.6 ENST00000684155.1 ENST00000376199.7 |
FOXP3
|
forkhead box P3 |
| chr6_+_50818857 | 0.05 |
ENST00000393655.4
|
TFAP2B
|
transcription factor AP-2 beta |
| chr5_-_95682968 | 0.05 |
ENST00000274432.13
|
SPATA9
|
spermatogenesis associated 9 |
| chr5_-_147782518 | 0.05 |
ENST00000507386.5
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2 |
| chr20_+_6007245 | 0.04 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr12_-_10807286 | 0.04 |
ENST00000240615.3
|
TAS2R8
|
taste 2 receptor member 8 |
| chr10_+_116591010 | 0.04 |
ENST00000530319.5
ENST00000527980.5 ENST00000471549.5 ENST00000534537.5 |
PNLIPRP1
|
pancreatic lipase related protein 1 |
| chr1_+_202416826 | 0.04 |
ENST00000466968.1
|
PPP1R12B
|
protein phosphatase 1 regulatory subunit 12B |
| chr5_+_157743703 | 0.04 |
ENST00000286307.6
|
LSM11
|
LSM11, U7 small nuclear RNA associated |
| chr3_+_174859315 | 0.03 |
ENST00000454872.6
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase like 2 |
| chr9_-_92482350 | 0.03 |
ENST00000375543.2
|
ASPN
|
asporin |
| chr17_-_40755328 | 0.03 |
ENST00000312150.5
|
KRT25
|
keratin 25 |
| chr12_-_51028234 | 0.03 |
ENST00000547688.7
ENST00000394904.9 |
SLC11A2
|
solute carrier family 11 member 2 |
| chr1_-_169630115 | 0.03 |
ENST00000263686.11
ENST00000367788.6 |
SELP
|
selectin P |
| chr5_+_173888335 | 0.03 |
ENST00000265085.10
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr2_-_177888728 | 0.03 |
ENST00000389683.7
|
PDE11A
|
phosphodiesterase 11A |
| chrM_+_10759 | 0.03 |
ENST00000361381.2
|
MT-ND4
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 4 |
| chr6_-_32763466 | 0.03 |
ENST00000427449.1
ENST00000411527.5 |
HLA-DQB2
|
major histocompatibility complex, class II, DQ beta 2 |
| chr20_+_45207025 | 0.03 |
ENST00000372781.4
|
SEMG1
|
semenogelin 1 |
| chr5_+_137889469 | 0.03 |
ENST00000290431.5
|
PKD2L2
|
polycystin 2 like 2, transient receptor potential cation channel |
| chrX_-_30577759 | 0.03 |
ENST00000378962.4
|
TASL
|
TLR adaptor interacting with endolysosomal SLC15A4 |
| chr13_-_98521998 | 0.02 |
ENST00000376547.7
|
STK24
|
serine/threonine kinase 24 |
| chrX_+_100584928 | 0.02 |
ENST00000373031.5
|
TNMD
|
tenomodulin |
| chr1_-_197067234 | 0.02 |
ENST00000367412.2
|
F13B
|
coagulation factor XIII B chain |
| chr22_-_32255344 | 0.02 |
ENST00000266086.6
|
SLC5A4
|
solute carrier family 5 member 4 |
| chr4_-_117085541 | 0.02 |
ENST00000310754.5
|
TRAM1L1
|
translocation associated membrane protein 1 like 1 |
| chr20_-_45972171 | 0.02 |
ENST00000322927.3
|
ZNF335
|
zinc finger protein 335 |
| chr11_-_14971179 | 0.01 |
ENST00000486207.5
|
CALCA
|
calcitonin related polypeptide alpha |
| chr10_-_32378720 | 0.01 |
ENST00000375110.6
|
EPC1
|
enhancer of polycomb homolog 1 |
| chr9_-_107489754 | 0.01 |
ENST00000610832.1
ENST00000374672.5 |
KLF4
|
Kruppel like factor 4 |
| chr18_+_74148508 | 0.01 |
ENST00000580087.5
ENST00000169551.11 |
TIMM21
|
translocase of inner mitochondrial membrane 21 |
| chr2_+_216498831 | 0.01 |
ENST00000491306.6
ENST00000600880.5 ENST00000446558.5 |
RPL37A
|
ribosomal protein L37a |
| chr6_-_48111132 | 0.00 |
ENST00000398738.3
ENST00000679966.1 ENST00000339488.9 |
PTCHD4
|
patched domain containing 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.1 | 0.5 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.1 | 0.9 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 0.3 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.1 | 0.3 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 0.3 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.1 | 0.2 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 | 0.6 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.1 | 0.2 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 | 0.3 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.2 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.0 | 0.4 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.1 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.6 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.1 | GO:2000690 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.1 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 0.0 | 0.1 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.1 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.0 | GO:0033092 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 | 3.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 2.7 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.0 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.0 | 0.1 | GO:0035604 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) |
| 0.0 | 1.3 | GO:0061045 | negative regulation of wound healing(GO:0061045) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.7 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.6 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.4 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0042008 | interleukin-33 binding(GO:0002113) interleukin-18 receptor activity(GO:0042008) |
| 0.1 | 0.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.2 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.1 | 0.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.2 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 1.7 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 0.6 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.3 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.0 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 0.0 | 0.4 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |