Project
Inflammatory response time course, HUVEC (Wada, 2009)
Navigation
Downloads
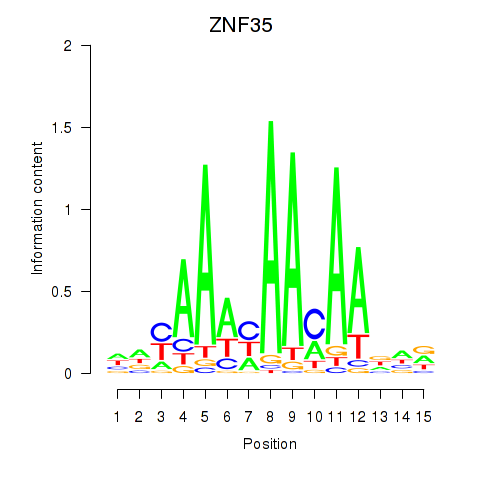
Results for ZNF35
Z-value: 0.33
Transcription factors associated with ZNF35
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF35
|
ENSG00000169981.11 | ZNF35 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF35 | hg38_v1_chr3_+_44648719_44648776 | -0.39 | 5.3e-02 | Click! |
Activity profile of ZNF35 motif
Sorted Z-values of ZNF35 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF35
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_61404076 | 1.24 |
ENST00000357977.5
|
NFIA
|
nuclear factor I A |
| chr7_-_28958321 | 0.95 |
ENST00000539664.3
|
TRIL
|
TLR4 interactor with leucine rich repeats |
| chr12_-_42483604 | 0.89 |
ENST00000640132.1
|
PRICKLE1
|
prickle planar cell polarity protein 1 |
| chr2_-_189580773 | 0.83 |
ENST00000261024.7
|
SLC40A1
|
solute carrier family 40 member 1 |
| chr1_+_84164962 | 0.75 |
ENST00000614872.4
ENST00000394839.6 |
PRKACB
|
protein kinase cAMP-activated catalytic subunit beta |
| chr2_+_66435558 | 0.60 |
ENST00000488550.5
|
MEIS1
|
Meis homeobox 1 |
| chr12_-_46372763 | 0.37 |
ENST00000256689.10
|
SLC38A2
|
solute carrier family 38 member 2 |
| chr9_+_128091436 | 0.36 |
ENST00000682371.1
ENST00000373066.9 ENST00000432073.6 |
SLC25A25
|
solute carrier family 25 member 25 |
| chr2_+_147845020 | 0.31 |
ENST00000241416.12
|
ACVR2A
|
activin A receptor type 2A |
| chr3_-_58537283 | 0.30 |
ENST00000459701.6
|
ACOX2
|
acyl-CoA oxidase 2 |
| chr3_-_58537181 | 0.29 |
ENST00000302819.10
|
ACOX2
|
acyl-CoA oxidase 2 |
| chr6_+_156776020 | 0.27 |
ENST00000346085.10
|
ARID1B
|
AT-rich interaction domain 1B |
| chr15_-_101252040 | 0.27 |
ENST00000254190.4
|
CHSY1
|
chondroitin sulfate synthase 1 |
| chr7_+_27242796 | 0.27 |
ENST00000496902.7
|
EVX1
|
even-skipped homeobox 1 |
| chr3_-_132684685 | 0.23 |
ENST00000512094.5
ENST00000632629.1 |
NPHP3
NPHP3-ACAD11
|
nephrocystin 3 NPHP3-ACAD11 readthrough (NMD candidate) |
| chr4_+_173370908 | 0.22 |
ENST00000296504.4
|
SAP30
|
Sin3A associated protein 30 |
| chr8_-_94262308 | 0.22 |
ENST00000297596.3
ENST00000396194.6 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr17_-_65561640 | 0.22 |
ENST00000618960.4
ENST00000307078.10 |
AXIN2
|
axin 2 |
| chr4_-_145938775 | 0.21 |
ENST00000508784.6
|
ZNF827
|
zinc finger protein 827 |
| chr18_+_41955186 | 0.21 |
ENST00000639914.1
ENST00000262039.9 ENST00000398870.7 ENST00000586545.5 ENST00000585528.5 |
PIK3C3
|
phosphatidylinositol 3-kinase catalytic subunit type 3 |
| chr17_-_445939 | 0.19 |
ENST00000329099.4
|
RFLNB
|
refilin B |
| chr4_-_83010709 | 0.18 |
ENST00000506560.5
ENST00000442461.6 ENST00000340417.8 ENST00000446851.6 |
LIN54
|
lin-54 DREAM MuvB core complex component |
| chr6_+_96563117 | 0.18 |
ENST00000450218.6
|
FHL5
|
four and a half LIM domains 5 |
| chr7_+_74657695 | 0.18 |
ENST00000573035.6
|
GTF2I
|
general transcription factor IIi |
| chr6_-_132763424 | 0.18 |
ENST00000532012.1
ENST00000525270.5 ENST00000530536.5 ENST00000524919.5 |
VNN2
|
vanin 2 |
| chr14_-_91732059 | 0.16 |
ENST00000553329.5
ENST00000256343.8 |
CATSPERB
|
cation channel sperm associated auxiliary subunit beta |
| chr19_+_926001 | 0.16 |
ENST00000263620.8
|
ARID3A
|
AT-rich interaction domain 3A |
| chr12_-_10939263 | 0.15 |
ENST00000537503.2
|
TAS2R14
|
taste 2 receptor member 14 |
| chr19_+_49591170 | 0.15 |
ENST00000418929.7
|
PRR12
|
proline rich 12 |
| chr8_-_144529048 | 0.14 |
ENST00000527462.1
ENST00000313465.5 ENST00000524821.6 |
C8orf82
|
chromosome 8 open reading frame 82 |
| chr10_+_61901678 | 0.14 |
ENST00000644638.1
ENST00000681100.1 ENST00000279873.12 |
ARID5B
|
AT-rich interaction domain 5B |
| chr7_+_27242700 | 0.14 |
ENST00000222761.7
|
EVX1
|
even-skipped homeobox 1 |
| chr20_+_58907981 | 0.13 |
ENST00000656419.1
|
GNAS
|
GNAS complex locus |
| chr7_+_80602150 | 0.12 |
ENST00000309881.11
|
CD36
|
CD36 molecule |
| chr22_-_36703723 | 0.11 |
ENST00000300105.7
|
CACNG2
|
calcium voltage-gated channel auxiliary subunit gamma 2 |
| chr13_-_35855627 | 0.11 |
ENST00000379893.5
|
DCLK1
|
doublecortin like kinase 1 |
| chr16_+_14833713 | 0.10 |
ENST00000287667.12
ENST00000620755.4 ENST00000610363.4 |
NOMO1
|
NODAL modulator 1 |
| chr19_-_14090695 | 0.10 |
ENST00000533683.7
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr14_-_31420531 | 0.09 |
ENST00000382464.6
ENST00000543095.7 |
HEATR5A
|
HEAT repeat containing 5A |
| chr4_+_74308463 | 0.09 |
ENST00000413830.6
|
EPGN
|
epithelial mitogen |
| chr12_+_93467506 | 0.09 |
ENST00000549982.6
ENST00000552217.6 ENST00000393128.8 ENST00000547098.5 |
MRPL42
|
mitochondrial ribosomal protein L42 |
| chr17_-_59151794 | 0.08 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr4_-_88823214 | 0.08 |
ENST00000513837.5
ENST00000503556.5 |
FAM13A
|
family with sequence similarity 13 member A |
| chr7_-_78771265 | 0.08 |
ENST00000630991.2
ENST00000629359.2 |
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr7_+_123848070 | 0.07 |
ENST00000476325.5
|
HYAL4
|
hyaluronidase 4 |
| chr12_-_102478539 | 0.06 |
ENST00000424202.6
|
IGF1
|
insulin like growth factor 1 |
| chr3_+_10115667 | 0.06 |
ENST00000530758.2
|
BRK1
|
BRICK1 subunit of SCAR/WAVE actin nucleating complex |
| chr5_-_39202991 | 0.05 |
ENST00000515010.5
|
FYB1
|
FYN binding protein 1 |
| chr16_+_76277568 | 0.05 |
ENST00000622250.4
|
CNTNAP4
|
contactin associated protein family member 4 |
| chr13_-_35855758 | 0.05 |
ENST00000615680.4
|
DCLK1
|
doublecortin like kinase 1 |
| chr14_-_73558945 | 0.03 |
ENST00000563329.1
ENST00000553558.6 ENST00000334988.2 |
HEATR4
|
HEAT repeat containing 4 |
| chr4_+_94455245 | 0.03 |
ENST00000508216.5
ENST00000514743.5 |
PDLIM5
|
PDZ and LIM domain 5 |
| chr3_+_69936583 | 0.03 |
ENST00000314557.10
ENST00000394351.9 |
MITF
|
melanocyte inducing transcription factor |
| chrM_+_4467 | 0.02 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH:ubiquinone oxidoreductase core subunit 2 |
| chr4_+_87832917 | 0.02 |
ENST00000395102.8
ENST00000497649.6 ENST00000540395.1 ENST00000560249.5 ENST00000511670.5 ENST00000361056.3 |
MEPE
|
matrix extracellular phosphoglycoprotein |
| chr4_-_98929092 | 0.02 |
ENST00000280892.10
ENST00000511644.5 ENST00000504432.5 ENST00000450253.7 ENST00000505992.1 |
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr13_+_77535681 | 0.01 |
ENST00000349847.4
|
SCEL
|
sciellin |
| chr13_+_77535669 | 0.01 |
ENST00000535157.5
|
SCEL
|
sciellin |
| chr3_-_64019334 | 0.01 |
ENST00000480205.5
|
PSMD6
|
proteasome 26S subunit, non-ATPase 6 |
| chr14_-_55191534 | 0.00 |
ENST00000395425.6
ENST00000247191.7 |
DLGAP5
|
DLG associated protein 5 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.2 | 0.9 | GO:2000691 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.1 | 0.4 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 | 0.7 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.2 | GO:0061182 | negative regulation of chondrocyte development(GO:0061182) |
| 0.1 | 0.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 1.2 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.4 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.3 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.0 | 0.6 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.6 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 0.4 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.0 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.9 | GO:0034142 | toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.0 | 0.1 | GO:2000334 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.1 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.2 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 0.2 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.3 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.1 | 0.8 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.3 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.4 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.9 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.8 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.7 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |