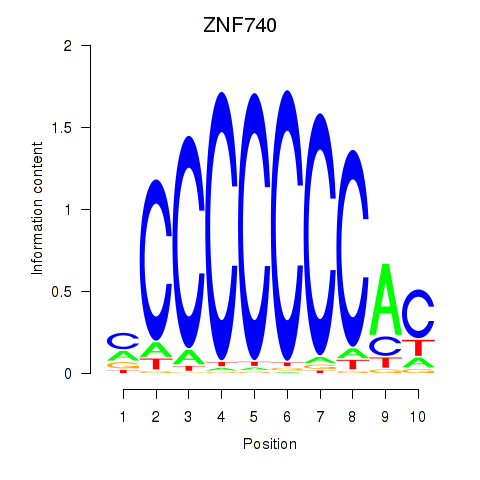
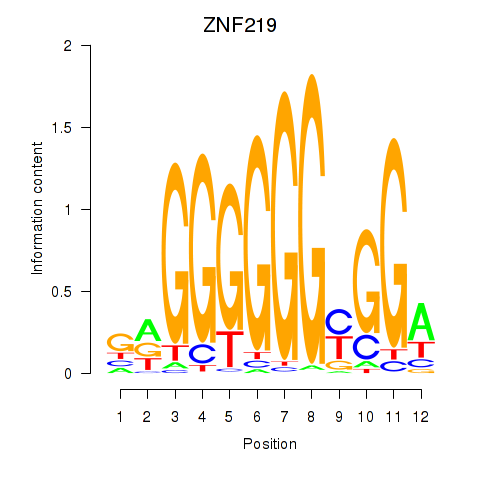
| 0.5 |
1.4 |
GO:1903422 |
negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.4 |
7.8 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.2 |
0.9 |
GO:0060708 |
spongiotrophoblast differentiation(GO:0060708) |
| 0.2 |
0.9 |
GO:0002296 |
T-helper 1 cell lineage commitment(GO:0002296) |
| 0.2 |
0.6 |
GO:0052026 |
modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.2 |
0.9 |
GO:2000325 |
regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.2 |
0.5 |
GO:2000740 |
amniotic stem cell differentiation(GO:0097086) negative regulation of dense core granule biogenesis(GO:2000706) negative regulation of mesenchymal stem cell differentiation(GO:2000740) regulation of amniotic stem cell differentiation(GO:2000797) negative regulation of amniotic stem cell differentiation(GO:2000798) |
| 0.2 |
0.9 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.2 |
0.7 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.2 |
1.0 |
GO:0003431 |
growth plate cartilage chondrocyte development(GO:0003431) |
| 0.2 |
1.2 |
GO:0060011 |
Sertoli cell proliferation(GO:0060011) |
| 0.2 |
2.0 |
GO:0000056 |
ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 |
0.5 |
GO:0060032 |
notochord regression(GO:0060032) |
| 0.2 |
0.7 |
GO:0030037 |
actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.2 |
0.5 |
GO:1903691 |
positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.2 |
0.5 |
GO:0008355 |
olfactory learning(GO:0008355) |
| 0.2 |
0.5 |
GO:0060128 |
corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.1 |
0.5 |
GO:1904800 |
negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.1 |
0.4 |
GO:0071464 |
cellular response to hydrostatic pressure(GO:0071464) |
| 0.1 |
1.6 |
GO:2000172 |
regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.1 |
0.6 |
GO:0006850 |
mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.1 |
1.5 |
GO:0051581 |
negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 |
0.4 |
GO:1901355 |
response to rapamycin(GO:1901355) |
| 0.1 |
0.4 |
GO:0001560 |
regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 |
0.3 |
GO:0043397 |
corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.1 |
0.3 |
GO:0002949 |
tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 |
0.7 |
GO:0098532 |
histone H3-K27 trimethylation(GO:0098532) |
| 0.1 |
0.2 |
GO:1990086 |
lens fiber cell apoptotic process(GO:1990086) |
| 0.1 |
0.4 |
GO:0035740 |
CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.1 |
0.7 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 |
0.4 |
GO:0072369 |
regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.1 |
1.1 |
GO:0019367 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 |
0.4 |
GO:0070889 |
mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 |
0.7 |
GO:1904100 |
regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.1 |
0.6 |
GO:0021966 |
corticospinal neuron axon guidance(GO:0021966) |
| 0.1 |
1.4 |
GO:0000338 |
protein deneddylation(GO:0000338) |
| 0.1 |
0.3 |
GO:1900194 |
negative regulation of oocyte maturation(GO:1900194) |
| 0.1 |
0.3 |
GO:1904328 |
regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.1 |
0.3 |
GO:0014873 |
response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.1 |
1.4 |
GO:0045636 |
positive regulation of melanocyte differentiation(GO:0045636) |
| 0.1 |
0.8 |
GO:0042048 |
olfactory behavior(GO:0042048) |
| 0.1 |
0.3 |
GO:0072275 |
metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 |
0.3 |
GO:0021897 |
forebrain astrocyte differentiation(GO:0021896) forebrain astrocyte development(GO:0021897) |
| 0.1 |
0.4 |
GO:0034721 |
histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.1 |
0.3 |
GO:0016103 |
diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 |
0.3 |
GO:0003064 |
regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.1 |
1.1 |
GO:0038129 |
directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.1 |
0.7 |
GO:0070164 |
negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 |
0.2 |
GO:2000591 |
positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.1 |
0.6 |
GO:0033183 |
negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.1 |
1.4 |
GO:1900017 |
positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 |
0.3 |
GO:2000638 |
regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 |
0.3 |
GO:1901165 |
positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 |
0.7 |
GO:0043353 |
enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 |
0.4 |
GO:0003219 |
cardiac right ventricle formation(GO:0003219) |
| 0.1 |
0.2 |
GO:1905006 |
negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) |
| 0.1 |
0.4 |
GO:0000430 |
regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.1 |
1.2 |
GO:0090394 |
negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.1 |
0.2 |
GO:1904637 |
response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) negative regulation of synapse maturation(GO:2000297) |
| 0.1 |
0.2 |
GO:0046338 |
phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 |
0.2 |
GO:0034414 |
tRNA 3'-trailer cleavage, endonucleolytic(GO:0034414) tRNA 3'-trailer cleavage(GO:0042779) |
| 0.1 |
0.5 |
GO:0003360 |
brainstem development(GO:0003360) |
| 0.1 |
0.2 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.1 |
0.2 |
GO:0035498 |
carnosine metabolic process(GO:0035498) |
| 0.1 |
0.2 |
GO:0003366 |
cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.1 |
0.4 |
GO:2000546 |
positive regulation of fat cell proliferation(GO:0070346) positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 |
0.9 |
GO:0097688 |
AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.1 |
1.5 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.1 |
0.3 |
GO:0061760 |
antifungal innate immune response(GO:0061760) |
| 0.1 |
0.3 |
GO:0035105 |
sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 |
0.4 |
GO:0001957 |
intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 |
0.2 |
GO:1990108 |
protein linear deubiquitination(GO:1990108) |
| 0.0 |
0.2 |
GO:0044805 |
late nucleophagy(GO:0044805) |
| 0.0 |
0.2 |
GO:0006344 |
maintenance of chromatin silencing(GO:0006344) |
| 0.0 |
0.5 |
GO:0010571 |
positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 |
0.1 |
GO:0014740 |
negative regulation of muscle hyperplasia(GO:0014740) |
| 0.0 |
0.3 |
GO:0001866 |
NK T cell proliferation(GO:0001866) |
| 0.0 |
0.3 |
GO:2000490 |
negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 |
2.8 |
GO:0043486 |
histone exchange(GO:0043486) |
| 0.0 |
0.4 |
GO:0034720 |
histone H3-K4 demethylation(GO:0034720) |
| 0.0 |
0.1 |
GO:0035565 |
regulation of pronephros size(GO:0035565) |
| 0.0 |
0.4 |
GO:0051725 |
protein de-ADP-ribosylation(GO:0051725) |
| 0.0 |
0.1 |
GO:0030910 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 |
0.2 |
GO:1904694 |
negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 |
0.1 |
GO:1903259 |
exon-exon junction complex disassembly(GO:1903259) |
| 0.0 |
0.8 |
GO:1904259 |
regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 |
0.9 |
GO:0010867 |
positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 |
0.2 |
GO:0022007 |
neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 |
0.5 |
GO:0006707 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
0.5 |
GO:1903025 |
regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 |
0.2 |
GO:1902775 |
mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 |
0.1 |
GO:0034148 |
regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) |
| 0.0 |
0.3 |
GO:0055005 |
ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 |
0.1 |
GO:0043376 |
regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 |
2.0 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.2 |
GO:0048633 |
positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 |
0.2 |
GO:0035513 |
oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 |
0.1 |
GO:0021520 |
spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 |
0.1 |
GO:0033385 |
geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 |
0.2 |
GO:0070172 |
positive regulation of tooth mineralization(GO:0070172) |
| 0.0 |
0.1 |
GO:0090309 |
positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 |
0.3 |
GO:0060478 |
acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 |
0.2 |
GO:0060373 |
regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 |
0.3 |
GO:0002051 |
osteoblast fate commitment(GO:0002051) |
| 0.0 |
0.0 |
GO:1902913 |
positive regulation of neuroepithelial cell differentiation(GO:1902913) |
| 0.0 |
0.3 |
GO:0061087 |
positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 |
0.5 |
GO:1901223 |
negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 |
0.2 |
GO:0031291 |
Ran protein signal transduction(GO:0031291) |
| 0.0 |
0.3 |
GO:0032000 |
positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 |
0.2 |
GO:2000676 |
positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 |
0.3 |
GO:0014877 |
response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 |
0.1 |
GO:0007538 |
primary sex determination(GO:0007538) |
| 0.0 |
0.4 |
GO:0046940 |
nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 |
0.1 |
GO:0072086 |
specification of loop of Henle identity(GO:0072086) |
| 0.0 |
0.4 |
GO:0072718 |
response to cisplatin(GO:0072718) |
| 0.0 |
1.3 |
GO:1900027 |
regulation of ruffle assembly(GO:1900027) |
| 0.0 |
0.1 |
GO:1904379 |
tail-anchored membrane protein insertion into ER membrane(GO:0071816) protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 |
0.1 |
GO:1905167 |
positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 |
0.1 |
GO:2001025 |
positive regulation of response to drug(GO:2001025) |
| 0.0 |
0.1 |
GO:0001880 |
Mullerian duct regression(GO:0001880) |
| 0.0 |
0.1 |
GO:1900248 |
cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 |
0.1 |
GO:0070105 |
positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 |
0.4 |
GO:0033235 |
positive regulation of protein sumoylation(GO:0033235) |
| 0.0 |
0.1 |
GO:0072367 |
regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 |
0.2 |
GO:0014050 |
negative regulation of glutamate secretion(GO:0014050) |
| 0.0 |
0.4 |
GO:0030157 |
pancreatic juice secretion(GO:0030157) |
| 0.0 |
0.1 |
GO:0048295 |
positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 |
0.4 |
GO:1902659 |
regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.0 |
0.1 |
GO:0032904 |
viral protein processing(GO:0019082) negative regulation of neurotrophin production(GO:0032900) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 |
0.1 |
GO:0003192 |
mitral valve formation(GO:0003192) |
| 0.0 |
0.2 |
GO:0007403 |
glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.0 |
0.2 |
GO:0048478 |
replication fork protection(GO:0048478) |
| 0.0 |
0.1 |
GO:0007499 |
ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 |
0.2 |
GO:0035865 |
cellular response to potassium ion(GO:0035865) |
| 0.0 |
0.2 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 |
1.7 |
GO:0051568 |
histone H3-K4 methylation(GO:0051568) |
| 0.0 |
0.1 |
GO:0070634 |
transepithelial ammonium transport(GO:0070634) |
| 0.0 |
0.2 |
GO:0032287 |
peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 |
0.4 |
GO:2000291 |
regulation of myoblast proliferation(GO:2000291) |
| 0.0 |
0.1 |
GO:0035674 |
tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 |
0.2 |
GO:0055059 |
asymmetric neuroblast division(GO:0055059) |
| 0.0 |
0.2 |
GO:2000489 |
regulation of hepatic stellate cell activation(GO:2000489) positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 |
1.5 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.0 |
0.6 |
GO:0048935 |
peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 |
0.3 |
GO:0060252 |
positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 |
0.1 |
GO:0014063 |
negative regulation of serotonin secretion(GO:0014063) |
| 0.0 |
0.6 |
GO:0021516 |
dorsal spinal cord development(GO:0021516) |
| 0.0 |
0.1 |
GO:0001922 |
B-1 B cell homeostasis(GO:0001922) |
| 0.0 |
0.1 |
GO:0033606 |
B cell receptor transport within lipid bilayer(GO:0032595) B cell receptor transport into membrane raft(GO:0032597) protein transport out of membrane raft(GO:0032599) chemokine receptor transport out of membrane raft(GO:0032600) negative regulation of transforming growth factor beta3 production(GO:0032913) chemokine receptor transport within lipid bilayer(GO:0033606) |
| 0.0 |
0.1 |
GO:0001831 |
trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 |
0.1 |
GO:0051461 |
regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 |
0.3 |
GO:0097107 |
postsynaptic density assembly(GO:0097107) gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 |
0.1 |
GO:0036269 |
swimming behavior(GO:0036269) |
| 0.0 |
0.4 |
GO:0043517 |
positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 |
0.1 |
GO:2001184 |
positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 |
0.1 |
GO:2001271 |
negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 |
0.4 |
GO:0007084 |
mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 |
0.4 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.0 |
0.3 |
GO:0014041 |
regulation of neuron maturation(GO:0014041) |
| 0.0 |
0.0 |
GO:0016539 |
intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 |
0.1 |
GO:0014057 |
positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 |
0.3 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.0 |
0.2 |
GO:0060052 |
neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 |
0.2 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 |
0.6 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 |
0.0 |
GO:1904154 |
positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 |
0.1 |
GO:2000543 |
positive regulation of gastrulation(GO:2000543) |
| 0.0 |
0.1 |
GO:0046833 |
positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 |
0.5 |
GO:0021511 |
spinal cord patterning(GO:0021511) |
| 0.0 |
0.1 |
GO:1900109 |
regulation of histone H3-K9 dimethylation(GO:1900109) positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.0 |
0.1 |
GO:0032483 |
regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 |
0.2 |
GO:0048664 |
neuron fate determination(GO:0048664) |
| 0.0 |
0.2 |
GO:0060267 |
positive regulation of respiratory burst(GO:0060267) |
| 0.0 |
0.6 |
GO:0031529 |
ruffle organization(GO:0031529) |
| 0.0 |
0.3 |
GO:0031643 |
positive regulation of myelination(GO:0031643) |
| 0.0 |
0.2 |
GO:0071394 |
cellular response to testosterone stimulus(GO:0071394) |
| 0.0 |
0.0 |
GO:0033078 |
extrathymic T cell differentiation(GO:0033078) regulation of extrathymic T cell differentiation(GO:0033082) sebum secreting cell proliferation(GO:1990654) |
| 0.0 |
0.1 |
GO:0021759 |
globus pallidus development(GO:0021759) |
| 0.0 |
0.3 |
GO:1900121 |
negative regulation of receptor binding(GO:1900121) |
| 0.0 |
0.1 |
GO:0090063 |
positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 |
0.2 |
GO:0035871 |
protein K11-linked deubiquitination(GO:0035871) |
| 0.0 |
0.3 |
GO:0042340 |
keratan sulfate catabolic process(GO:0042340) |
| 0.0 |
0.1 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 |
0.1 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 |
0.1 |
GO:0010792 |
DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 |
0.3 |
GO:0032486 |
Rap protein signal transduction(GO:0032486) |
| 0.0 |
0.1 |
GO:0035469 |
determination of pancreatic left/right asymmetry(GO:0035469) determination of liver left/right asymmetry(GO:0071910) |
| 0.0 |
0.1 |
GO:0060158 |
phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 |
1.0 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 |
0.1 |
GO:0006499 |
N-terminal protein myristoylation(GO:0006499) |
| 0.0 |
0.2 |
GO:1904152 |
regulation of retrograde protein transport, ER to cytosol(GO:1904152) |
| 0.0 |
0.1 |
GO:0015742 |
alpha-ketoglutarate transport(GO:0015742) |
| 0.0 |
0.1 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.0 |
0.2 |
GO:0097091 |
synaptic vesicle clustering(GO:0097091) |
| 0.0 |
0.2 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 |
0.1 |
GO:0019509 |
L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 |
0.1 |
GO:0061484 |
hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 |
0.3 |
GO:0032968 |
positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 |
0.2 |
GO:0070544 |
histone H3-K36 demethylation(GO:0070544) |
| 0.0 |
0.3 |
GO:0001829 |
trophectodermal cell differentiation(GO:0001829) |
| 0.0 |
0.4 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.0 |
0.0 |
GO:0061026 |
central nervous system morphogenesis(GO:0021551) cardiac muscle tissue regeneration(GO:0061026) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.0 |
0.1 |
GO:0033211 |
adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 |
0.3 |
GO:0032354 |
response to follicle-stimulating hormone(GO:0032354) |
| 0.0 |
0.2 |
GO:0010944 |
negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 |
0.1 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 |
1.7 |
GO:0032436 |
positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 |
0.3 |
GO:0060732 |
positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 |
0.1 |
GO:0051684 |
maintenance of Golgi location(GO:0051684) |
| 0.0 |
0.1 |
GO:0060576 |
intestinal epithelial cell development(GO:0060576) |
| 0.0 |
0.1 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 |
0.6 |
GO:0033120 |
positive regulation of RNA splicing(GO:0033120) |
| 0.0 |
1.3 |
GO:0006521 |
regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 |
0.1 |
GO:0030917 |
midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 |
0.3 |
GO:0018023 |
peptidyl-lysine trimethylation(GO:0018023) |
| 0.0 |
0.1 |
GO:0000160 |
phosphorelay signal transduction system(GO:0000160) |
| 0.0 |
0.1 |
GO:0007512 |
adult heart development(GO:0007512) |
| 0.0 |
0.2 |
GO:0002091 |
negative regulation of receptor internalization(GO:0002091) |
| 0.0 |
0.4 |
GO:2001222 |
regulation of neuron migration(GO:2001222) |
| 0.0 |
0.2 |
GO:0045721 |
negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 |
0.2 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 |
0.2 |
GO:0006337 |
nucleosome disassembly(GO:0006337) |
| 0.0 |
0.1 |
GO:0021979 |
hypothalamus cell differentiation(GO:0021979) |
| 0.0 |
0.1 |
GO:0070127 |
tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 |
0.5 |
GO:0043403 |
skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 |
0.4 |
GO:0045736 |
negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) |
| 0.0 |
0.9 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.0 |
0.2 |
GO:0030889 |
negative regulation of B cell proliferation(GO:0030889) |
| 0.0 |
0.2 |
GO:0045955 |
negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.0 |
0.2 |
GO:0015732 |
prostaglandin transport(GO:0015732) |
| 0.0 |
0.2 |
GO:0032196 |
transposition(GO:0032196) |
| 0.0 |
0.2 |
GO:0043562 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 |
0.0 |
GO:1901842 |
negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 |
0.2 |
GO:0090360 |
platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 |
1.6 |
GO:0043484 |
regulation of RNA splicing(GO:0043484) |