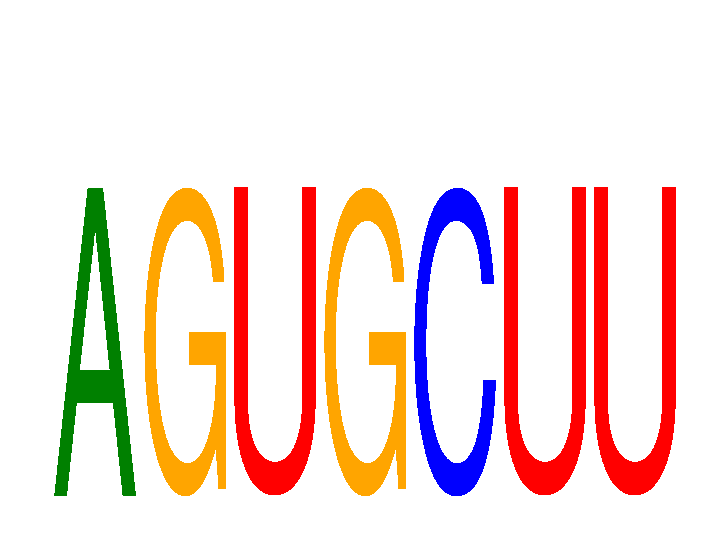Project
Illumina Body Map 2 (GSE30611)
Navigation
Downloads
Results for AGUGCUU
Z-value: 0.73
miRNA associated with seed AGUGCUU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-302c-3p.2
|
|
|
hsa-miR-520f-3p
|
MIMAT0002830 |
Activity profile of AGUGCUU motif
Sorted Z-values of AGUGCUU motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_28177102 | 1.86 |
ENST00000413878.2
ENST00000269141.8 |
CDH2
|
cadherin 2 |
| chr8_+_78516329 | 1.73 |
ENST00000396418.7
ENST00000352966.9 |
PKIA
|
cAMP-dependent protein kinase inhibitor alpha |
| chr9_+_2621766 | 1.37 |
ENST00000382100.8
|
VLDLR
|
very low density lipoprotein receptor |
| chr9_-_109167159 | 1.32 |
ENST00000561981.5
|
FRRS1L
|
ferric chelate reductase 1 like |
| chr4_-_52659238 | 1.21 |
ENST00000451218.6
ENST00000441222.8 |
USP46
|
ubiquitin specific peptidase 46 |
| chr1_-_225941383 | 1.17 |
ENST00000420304.6
|
LEFTY2
|
left-right determination factor 2 |
| chr10_-_59709842 | 1.15 |
ENST00000395348.8
|
SLC16A9
|
solute carrier family 16 member 9 |
| chrX_+_106168297 | 1.06 |
ENST00000337685.6
ENST00000357175.6 |
PWWP3B
|
PWWP domain containing 3B |
| chr20_-_49482645 | 1.03 |
ENST00000371741.6
|
KCNB1
|
potassium voltage-gated channel subfamily B member 1 |
| chr20_+_9514562 | 0.99 |
ENST00000246070.3
|
LAMP5
|
lysosomal associated membrane protein family member 5 |
| chr15_-_78234513 | 0.97 |
ENST00000558130.1
ENST00000258873.9 |
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr15_-_73368951 | 0.97 |
ENST00000261917.4
|
HCN4
|
hyperpolarization activated cyclic nucleotide gated potassium channel 4 |
| chr2_+_11534039 | 0.97 |
ENST00000381486.7
|
GREB1
|
growth regulating estrogen receptor binding 1 |
| chr1_-_225889143 | 0.90 |
ENST00000272134.5
|
LEFTY1
|
left-right determination factor 1 |
| chr3_+_119294337 | 0.83 |
ENST00000264245.9
|
ARHGAP31
|
Rho GTPase activating protein 31 |
| chr12_-_31591129 | 0.78 |
ENST00000389082.10
|
DENND5B
|
DENN domain containing 5B |
| chr17_-_17972374 | 0.78 |
ENST00000318094.14
ENST00000540946.5 ENST00000379504.8 ENST00000542206.5 ENST00000395739.8 ENST00000581396.5 ENST00000535933.5 ENST00000579586.1 |
TOM1L2
|
target of myb1 like 2 membrane trafficking protein |
| chr6_+_17281341 | 0.74 |
ENST00000379052.10
|
RBM24
|
RNA binding motif protein 24 |
| chrX_+_28587411 | 0.69 |
ENST00000378993.6
|
IL1RAPL1
|
interleukin 1 receptor accessory protein like 1 |
| chr17_+_4583998 | 0.66 |
ENST00000338859.8
|
SMTNL2
|
smoothelin like 2 |
| chrX_-_63755032 | 0.66 |
ENST00000624538.2
ENST00000636276.1 ENST00000624843.3 ENST00000671907.1 ENST00000624210.3 ENST00000374870.8 ENST00000635967.1 ENST00000253401.10 ENST00000672194.1 ENST00000637723.2 ENST00000637417.1 ENST00000637520.1 ENST00000374872.4 ENST00000636926.1 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor 9 |
| chr14_-_34713788 | 0.65 |
ENST00000341223.8
|
CFL2
|
cofilin 2 |
| chr17_+_72121012 | 0.63 |
ENST00000245479.3
|
SOX9
|
SRY-box transcription factor 9 |
| chr13_-_25172278 | 0.62 |
ENST00000515384.2
ENST00000357816.2 |
AMER2
|
APC membrane recruitment protein 2 |
| chr6_-_89412219 | 0.60 |
ENST00000369415.9
|
RRAGD
|
Ras related GTP binding D |
| chr7_+_107891135 | 0.58 |
ENST00000639772.1
ENST00000440410.5 ENST00000437604.6 ENST00000205402.10 |
DLD
|
dihydrolipoamide dehydrogenase |
| chr1_-_23369813 | 0.58 |
ENST00000314011.9
|
ZNF436
|
zinc finger protein 436 |
| chr2_-_212538766 | 0.57 |
ENST00000342788.9
|
ERBB4
|
erb-b2 receptor tyrosine kinase 4 |
| chr1_-_230426293 | 0.56 |
ENST00000391860.7
|
PGBD5
|
piggyBac transposable element derived 5 |
| chr11_-_27472698 | 0.56 |
ENST00000389858.4
ENST00000379214.9 |
LGR4
|
leucine rich repeat containing G protein-coupled receptor 4 |
| chr1_+_20186076 | 0.56 |
ENST00000375099.4
|
UBXN10
|
UBX domain protein 10 |
| chr19_-_31349408 | 0.55 |
ENST00000240587.5
|
TSHZ3
|
teashirt zinc finger homeobox 3 |
| chr1_-_84690406 | 0.55 |
ENST00000605755.5
ENST00000342203.8 ENST00000437941.6 |
SSX2IP
|
SSX family member 2 interacting protein |
| chr8_-_104588998 | 0.55 |
ENST00000424843.6
|
LRP12
|
LDL receptor related protein 12 |
| chrX_+_111096136 | 0.54 |
ENST00000372007.10
|
PAK3
|
p21 (RAC1) activated kinase 3 |
| chr15_-_93073111 | 0.54 |
ENST00000557420.1
ENST00000542321.6 |
RGMA
|
repulsive guidance molecule BMP co-receptor a |
| chr1_-_155562693 | 0.54 |
ENST00000368346.7
ENST00000392403.8 ENST00000679333.1 ENST00000679133.1 |
ASH1L
|
ASH1 like histone lysine methyltransferase |
| chr9_+_106863121 | 0.53 |
ENST00000472574.1
ENST00000277225.10 |
ZNF462
|
zinc finger protein 462 |
| chr11_+_9384621 | 0.52 |
ENST00000379719.8
ENST00000527431.1 ENST00000630083.1 |
IPO7
|
importin 7 |
| chr11_+_61752603 | 0.51 |
ENST00000278836.10
|
MYRF
|
myelin regulatory factor |
| chr9_-_10612966 | 0.50 |
ENST00000381196.9
|
PTPRD
|
protein tyrosine phosphatase receptor type D |
| chrX_-_24027186 | 0.50 |
ENST00000328046.8
|
KLHL15
|
kelch like family member 15 |
| chr12_+_4273751 | 0.49 |
ENST00000675880.1
ENST00000261254.8 |
CCND2
|
cyclin D2 |
| chr3_+_156674579 | 0.47 |
ENST00000295924.12
|
TIPARP
|
TCDD inducible poly(ADP-ribose) polymerase |
| chr16_+_25691953 | 0.47 |
ENST00000331351.6
|
HS3ST4
|
heparan sulfate-glucosamine 3-sulfotransferase 4 |
| chr2_+_29115367 | 0.46 |
ENST00000320081.10
|
CLIP4
|
CAP-Gly domain containing linker protein family member 4 |
| chr1_+_11980181 | 0.45 |
ENST00000444836.5
ENST00000674817.1 ENST00000675053.1 ENST00000675817.1 ENST00000675298.1 ENST00000676369.1 ENST00000412236.2 ENST00000675530.1 ENST00000674548.1 ENST00000674658.1 ENST00000674910.1 ENST00000675231.1 |
MFN2
|
mitofusin 2 |
| chr1_+_42380772 | 0.45 |
ENST00000431473.4
ENST00000410070.6 |
RIMKLA
|
ribosomal modification protein rimK like family member A |
| chr11_-_106022209 | 0.45 |
ENST00000301919.9
ENST00000534458.1 ENST00000530108.1 ENST00000530788.1 |
MSANTD4
|
Myb/SANT DNA binding domain containing 4 with coiled-coils |
| chr3_-_165196369 | 0.44 |
ENST00000475390.2
|
SLITRK3
|
SLIT and NTRK like family member 3 |
| chr2_+_99337364 | 0.44 |
ENST00000617677.1
ENST00000289371.11 |
EIF5B
|
eukaryotic translation initiation factor 5B |
| chr1_+_203626775 | 0.44 |
ENST00000367218.7
|
ATP2B4
|
ATPase plasma membrane Ca2+ transporting 4 |
| chr11_-_103092145 | 0.41 |
ENST00000260247.10
ENST00000531543.1 |
DCUN1D5
|
defective in cullin neddylation 1 domain containing 5 |
| chr6_-_99349647 | 0.41 |
ENST00000389677.6
|
FAXC
|
failed axon connections homolog, metaxin like GST domain containing |
| chr3_-_45995807 | 0.40 |
ENST00000535325.5
ENST00000296137.7 |
FYCO1
|
FYVE and coiled-coil domain autophagy adaptor 1 |
| chr12_+_20368495 | 0.40 |
ENST00000359062.4
|
PDE3A
|
phosphodiesterase 3A |
| chr4_+_183905266 | 0.39 |
ENST00000308497.9
|
STOX2
|
storkhead box 2 |
| chr10_+_68560317 | 0.38 |
ENST00000373644.5
|
TET1
|
tet methylcytosine dioxygenase 1 |
| chr11_+_35662739 | 0.38 |
ENST00000299413.7
|
TRIM44
|
tripartite motif containing 44 |
| chr9_+_125747345 | 0.38 |
ENST00000342287.9
ENST00000373489.10 ENST00000373487.8 |
PBX3
|
PBX homeobox 3 |
| chr14_-_53152371 | 0.38 |
ENST00000323669.10
|
DDHD1
|
DDHD domain containing 1 |
| chr8_+_105318428 | 0.38 |
ENST00000407775.7
|
ZFPM2
|
zinc finger protein, FOG family member 2 |
| chr12_+_109713817 | 0.37 |
ENST00000538780.2
|
FAM222A
|
family with sequence similarity 222 member A |
| chr6_-_89118002 | 0.36 |
ENST00000452027.3
|
SRSF12
|
serine and arginine rich splicing factor 12 |
| chr3_+_151086889 | 0.36 |
ENST00000474524.5
ENST00000273432.8 |
MED12L
|
mediator complex subunit 12L |
| chr1_-_174022339 | 0.36 |
ENST00000367696.7
|
RC3H1
|
ring finger and CCCH-type domains 1 |
| chr6_+_24667026 | 0.36 |
ENST00000537591.5
ENST00000230048.5 |
ACOT13
|
acyl-CoA thioesterase 13 |
| chrX_-_20116871 | 0.36 |
ENST00000379651.7
ENST00000443379.7 ENST00000379643.10 |
MAP7D2
|
MAP7 domain containing 2 |
| chr10_+_89392546 | 0.34 |
ENST00000546318.2
ENST00000371804.4 |
IFIT1
|
interferon induced protein with tetratricopeptide repeats 1 |
| chr2_+_169827432 | 0.34 |
ENST00000272793.11
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr13_-_78659124 | 0.34 |
ENST00000282003.7
|
OBI1
|
ORC ubiquitin ligase 1 |
| chr3_-_55489938 | 0.34 |
ENST00000474267.5
|
WNT5A
|
Wnt family member 5A |
| chr22_-_35840218 | 0.34 |
ENST00000414461.6
ENST00000416721.6 ENST00000449924.6 ENST00000262829.11 ENST00000397305.3 |
RBFOX2
|
RNA binding fox-1 homolog 2 |
| chr5_-_45696326 | 0.34 |
ENST00000673735.1
ENST00000303230.6 |
HCN1
|
hyperpolarization activated cyclic nucleotide gated potassium channel 1 |
| chr16_-_19522062 | 0.34 |
ENST00000353258.8
|
GDE1
|
glycerophosphodiester phosphodiesterase 1 |
| chr4_+_133149278 | 0.33 |
ENST00000264360.7
|
PCDH10
|
protocadherin 10 |
| chr5_-_138033021 | 0.33 |
ENST00000033079.7
|
FAM13B
|
family with sequence similarity 13 member B |
| chr1_-_156082412 | 0.33 |
ENST00000532414.3
|
MEX3A
|
mex-3 RNA binding family member A |
| chr6_+_57172290 | 0.33 |
ENST00000370693.5
|
BAG2
|
BAG cochaperone 2 |
| chr5_-_172006817 | 0.32 |
ENST00000296933.10
|
FBXW11
|
F-box and WD repeat domain containing 11 |
| chr22_-_21867610 | 0.32 |
ENST00000215832.11
ENST00000398822.7 |
MAPK1
|
mitogen-activated protein kinase 1 |
| chr22_+_29073024 | 0.32 |
ENST00000400335.9
|
KREMEN1
|
kringle containing transmembrane protein 1 |
| chr21_-_39313610 | 0.32 |
ENST00000342449.8
ENST00000341322.4 |
BRWD1
|
bromodomain and WD repeat domain containing 1 |
| chr18_-_75209126 | 0.31 |
ENST00000322342.4
|
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2 |
| chr10_+_113854610 | 0.31 |
ENST00000369301.3
|
NHLRC2
|
NHL repeat containing 2 |
| chr11_-_77820706 | 0.31 |
ENST00000440064.2
ENST00000528095.5 ENST00000308488.11 |
RSF1
|
remodeling and spacing factor 1 |
| chr3_+_179347686 | 0.31 |
ENST00000471841.6
|
MFN1
|
mitofusin 1 |
| chr16_+_24256313 | 0.30 |
ENST00000005284.4
|
CACNG3
|
calcium voltage-gated channel auxiliary subunit gamma 3 |
| chr12_+_55743110 | 0.30 |
ENST00000257868.10
|
GDF11
|
growth differentiation factor 11 |
| chr17_-_1179940 | 0.30 |
ENST00000302538.10
|
ABR
|
ABR activator of RhoGEF and GTPase |
| chr1_-_20486197 | 0.30 |
ENST00000375078.4
|
CAMK2N1
|
calcium/calmodulin dependent protein kinase II inhibitor 1 |
| chr10_+_100535927 | 0.30 |
ENST00000299163.7
|
HIF1AN
|
hypoxia inducible factor 1 subunit alpha inhibitor |
| chr6_+_151690492 | 0.30 |
ENST00000404742.5
ENST00000440973.5 |
ESR1
|
estrogen receptor 1 |
| chr1_+_214603173 | 0.29 |
ENST00000366955.8
|
CENPF
|
centromere protein F |
| chr16_+_7332839 | 0.29 |
ENST00000355637.9
|
RBFOX1
|
RNA binding fox-1 homolog 1 |
| chr7_+_129434424 | 0.28 |
ENST00000249344.7
ENST00000435494.2 |
STRIP2
|
striatin interacting protein 2 |
| chr5_+_98769273 | 0.28 |
ENST00000308234.11
|
RGMB
|
repulsive guidance molecule BMP co-receptor b |
| chr17_+_29390326 | 0.28 |
ENST00000261716.8
|
TAOK1
|
TAO kinase 1 |
| chr8_+_57994455 | 0.27 |
ENST00000361488.7
|
FAM110B
|
family with sequence similarity 110 member B |
| chr9_+_126805003 | 0.27 |
ENST00000449886.5
ENST00000450858.1 ENST00000373464.5 |
ZBTB43
|
zinc finger and BTB domain containing 43 |
| chr1_-_101996919 | 0.26 |
ENST00000370103.9
|
OLFM3
|
olfactomedin 3 |
| chr1_-_77682639 | 0.26 |
ENST00000370801.8
ENST00000433749.5 |
ZZZ3
|
zinc finger ZZ-type containing 3 |
| chr3_-_180036918 | 0.26 |
ENST00000465751.5
ENST00000467460.6 ENST00000472994.5 |
PEX5L
|
peroxisomal biogenesis factor 5 like |
| chr8_-_118621901 | 0.26 |
ENST00000409003.5
ENST00000524796.6 ENST00000314727.9 |
SAMD12
|
sterile alpha motif domain containing 12 |
| chr11_-_119729158 | 0.25 |
ENST00000264025.8
|
NECTIN1
|
nectin cell adhesion molecule 1 |
| chr1_+_220528112 | 0.25 |
ENST00000366917.6
ENST00000402574.5 ENST00000611084.4 ENST00000366918.8 |
MARK1
|
microtubule affinity regulating kinase 1 |
| chr20_-_43189733 | 0.25 |
ENST00000373187.5
ENST00000356100.6 ENST00000373184.5 ENST00000373190.5 |
PTPRT
|
protein tyrosine phosphatase receptor type T |
| chr5_-_32444722 | 0.25 |
ENST00000265069.13
|
ZFR
|
zinc finger RNA binding protein |
| chr4_+_6269831 | 0.25 |
ENST00000503569.5
ENST00000673991.1 ENST00000682275.1 ENST00000226760.5 |
WFS1
|
wolframin ER transmembrane glycoprotein |
| chr4_-_42657085 | 0.25 |
ENST00000264449.14
ENST00000510289.1 ENST00000381668.9 |
ATP8A1
|
ATPase phospholipid transporting 8A1 |
| chr20_+_19212624 | 0.25 |
ENST00000328041.11
|
SLC24A3
|
solute carrier family 24 member 3 |
| chr10_-_27240743 | 0.25 |
ENST00000677901.1
ENST00000677960.1 ENST00000677440.1 ENST00000396271.8 ENST00000677141.1 ENST00000677311.1 ENST00000677667.1 ENST00000677200.1 ENST00000676997.1 ENST00000676511.1 |
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr1_+_107056656 | 0.25 |
ENST00000370078.2
|
PRMT6
|
protein arginine methyltransferase 6 |
| chr6_+_118894144 | 0.24 |
ENST00000229595.6
|
ASF1A
|
anti-silencing function 1A histone chaperone |
| chr2_+_197515565 | 0.24 |
ENST00000233892.8
ENST00000409916.5 |
MOB4
|
MOB family member 4, phocein |
| chr1_+_111619751 | 0.24 |
ENST00000433097.5
ENST00000369709.3 |
RAP1A
|
RAP1A, member of RAS oncogene family |
| chr7_+_87345656 | 0.24 |
ENST00000331536.8
ENST00000419147.6 ENST00000412227.6 |
CROT
|
carnitine O-octanoyltransferase |
| chr1_+_184386978 | 0.24 |
ENST00000235307.7
|
C1orf21
|
chromosome 1 open reading frame 21 |
| chr10_+_91220603 | 0.23 |
ENST00000336126.6
|
PCGF5
|
polycomb group ring finger 5 |
| chr17_+_5078450 | 0.23 |
ENST00000318833.4
|
ZFP3
|
ZFP3 zinc finger protein |
| chr9_-_16870662 | 0.23 |
ENST00000380672.9
|
BNC2
|
basonuclin 2 |
| chr7_-_103149182 | 0.23 |
ENST00000417955.5
ENST00000341533.8 ENST00000425379.1 |
NAPEPLD
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr10_+_84328625 | 0.22 |
ENST00000224756.12
|
CCSER2
|
coiled-coil serine rich protein 2 |
| chr10_+_1049476 | 0.22 |
ENST00000358220.5
|
WDR37
|
WD repeat domain 37 |
| chr16_-_85688912 | 0.22 |
ENST00000253462.8
|
GINS2
|
GINS complex subunit 2 |
| chr2_+_149330506 | 0.22 |
ENST00000334166.9
|
LYPD6
|
LY6/PLAUR domain containing 6 |
| chr20_+_58309704 | 0.22 |
ENST00000244040.4
|
RAB22A
|
RAB22A, member RAS oncogene family |
| chr16_-_66696680 | 0.22 |
ENST00000330687.8
ENST00000563952.1 ENST00000394106.7 |
CMTM4
|
CKLF like MARVEL transmembrane domain containing 4 |
| chr1_+_153658687 | 0.22 |
ENST00000368685.6
|
SNAPIN
|
SNAP associated protein |
| chr6_-_116060859 | 0.22 |
ENST00000606080.2
|
FRK
|
fyn related Src family tyrosine kinase |
| chr15_-_43824675 | 0.22 |
ENST00000267812.4
|
MFAP1
|
microfibril associated protein 1 |
| chrX_-_72714278 | 0.22 |
ENST00000373542.9
ENST00000373545.7 |
PHKA1
|
phosphorylase kinase regulatory subunit alpha 1 |
| chr2_+_173354820 | 0.22 |
ENST00000347703.7
ENST00000410101.7 ENST00000410019.3 ENST00000306721.8 |
CDCA7
|
cell division cycle associated 7 |
| chr6_-_145814744 | 0.22 |
ENST00000237281.5
|
FBXO30
|
F-box protein 30 |
| chr17_-_80476597 | 0.21 |
ENST00000306773.5
|
NPTX1
|
neuronal pentraxin 1 |
| chr2_-_217944005 | 0.21 |
ENST00000611415.4
ENST00000615025.5 ENST00000449814.1 ENST00000171887.8 |
TNS1
|
tensin 1 |
| chr2_-_183038405 | 0.21 |
ENST00000361354.9
|
NCKAP1
|
NCK associated protein 1 |
| chr11_+_22666604 | 0.21 |
ENST00000454584.6
|
GAS2
|
growth arrest specific 2 |
| chr3_+_25428233 | 0.20 |
ENST00000437042.6
ENST00000330688.9 |
RARB
|
retinoic acid receptor beta |
| chr16_-_73048104 | 0.20 |
ENST00000268489.10
|
ZFHX3
|
zinc finger homeobox 3 |
| chr11_+_24497155 | 0.20 |
ENST00000529015.5
ENST00000533227.5 |
LUZP2
|
leucine zipper protein 2 |
| chr22_-_40856565 | 0.20 |
ENST00000620312.4
ENST00000216218.8 |
ST13
|
ST13 Hsp70 interacting protein |
| chr1_+_3690654 | 0.19 |
ENST00000378285.5
ENST00000378280.5 ENST00000378288.8 |
TP73
|
tumor protein p73 |
| chr2_-_165794190 | 0.19 |
ENST00000392701.8
ENST00000422973.1 |
GALNT3
|
polypeptide N-acetylgalactosaminyltransferase 3 |
| chr6_-_89638716 | 0.19 |
ENST00000626778.2
ENST00000520441.5 ENST00000523377.2 ENST00000520318.1 |
LYRM2
|
LYR motif containing 2 |
| chr6_-_56851888 | 0.19 |
ENST00000312431.10
ENST00000520645.5 |
DST
|
dystonin |
| chr2_-_69643703 | 0.19 |
ENST00000406297.7
ENST00000409085.9 |
AAK1
|
AP2 associated kinase 1 |
| chr16_-_3443446 | 0.18 |
ENST00000301744.7
|
ZNF597
|
zinc finger protein 597 |
| chr6_+_13615322 | 0.18 |
ENST00000451315.7
|
NOL7
|
nucleolar protein 7 |
| chr10_+_117542416 | 0.18 |
ENST00000442245.5
|
EMX2
|
empty spiracles homeobox 2 |
| chr9_-_122931477 | 0.18 |
ENST00000373656.4
|
ZBTB26
|
zinc finger and BTB domain containing 26 |
| chr2_+_26034069 | 0.18 |
ENST00000264710.5
|
RAB10
|
RAB10, member RAS oncogene family |
| chrX_+_129540236 | 0.18 |
ENST00000371113.9
ENST00000357121.5 |
OCRL
|
OCRL inositol polyphosphate-5-phosphatase |
| chrX_+_24054931 | 0.17 |
ENST00000253039.9
ENST00000423068.1 |
EIF2S3
|
eukaryotic translation initiation factor 2 subunit gamma |
| chr15_-_65377991 | 0.17 |
ENST00000327987.9
|
IGDCC3
|
immunoglobulin superfamily DCC subclass member 3 |
| chr1_+_32741779 | 0.17 |
ENST00000401073.7
|
KIAA1522
|
KIAA1522 |
| chr10_+_84139491 | 0.17 |
ENST00000372134.6
|
GHITM
|
growth hormone inducible transmembrane protein |
| chr8_-_102655707 | 0.17 |
ENST00000285407.11
|
KLF10
|
Kruppel like factor 10 |
| chr3_+_132417487 | 0.17 |
ENST00000260818.11
|
DNAJC13
|
DnaJ heat shock protein family (Hsp40) member C13 |
| chr7_+_100015588 | 0.17 |
ENST00000324306.11
ENST00000426572.5 |
ZKSCAN1
|
zinc finger with KRAB and SCAN domains 1 |
| chr8_-_52714414 | 0.17 |
ENST00000435644.6
ENST00000518710.5 ENST00000025008.10 ENST00000517963.1 |
RB1CC1
|
RB1 inducible coiled-coil 1 |
| chr1_-_70205531 | 0.17 |
ENST00000370952.4
|
LRRC40
|
leucine rich repeat containing 40 |
| chr14_+_52552830 | 0.17 |
ENST00000321662.11
|
GPR137C
|
G protein-coupled receptor 137C |
| chr13_-_99971739 | 0.17 |
ENST00000267294.4
|
ZIC5
|
Zic family member 5 |
| chr17_+_70169516 | 0.17 |
ENST00000243457.4
|
KCNJ2
|
potassium inwardly rectifying channel subfamily J member 2 |
| chr10_-_59906509 | 0.17 |
ENST00000263102.7
|
CCDC6
|
coiled-coil domain containing 6 |
| chr2_+_177392734 | 0.16 |
ENST00000680770.1
ENST00000637633.2 ENST00000679459.1 ENST00000409888.1 ENST00000264167.11 ENST00000642466.2 |
AGPS
|
alkylglycerone phosphate synthase |
| chr2_+_174334947 | 0.16 |
ENST00000394967.3
|
SP9
|
Sp9 transcription factor |
| chr6_-_42451910 | 0.16 |
ENST00000372922.8
ENST00000541110.5 |
TRERF1
|
transcriptional regulating factor 1 |
| chrX_-_84188148 | 0.16 |
ENST00000262752.5
|
RPS6KA6
|
ribosomal protein S6 kinase A6 |
| chr6_-_157323498 | 0.16 |
ENST00000400788.9
ENST00000367144.4 |
TMEM242
|
transmembrane protein 242 |
| chr1_+_88684222 | 0.16 |
ENST00000316005.11
ENST00000370521.8 |
PKN2
|
protein kinase N2 |
| chr16_-_47461259 | 0.16 |
ENST00000544001.6
|
ITFG1
|
integrin alpha FG-GAP repeat containing 1 |
| chr2_+_6917404 | 0.16 |
ENST00000320892.11
|
RNF144A
|
ring finger protein 144A |
| chr2_-_24971900 | 0.15 |
ENST00000264711.7
|
DNAJC27
|
DnaJ heat shock protein family (Hsp40) member C27 |
| chr8_+_81732434 | 0.15 |
ENST00000297265.5
|
CHMP4C
|
charged multivesicular body protein 4C |
| chr3_+_84958963 | 0.15 |
ENST00000383699.8
|
CADM2
|
cell adhesion molecule 2 |
| chr2_-_86721122 | 0.15 |
ENST00000604011.5
|
RNF103-CHMP3
|
RNF103-CHMP3 readthrough |
| chr6_+_87155537 | 0.15 |
ENST00000369577.8
ENST00000518845.1 ENST00000339907.8 ENST00000496806.2 |
ZNF292
|
zinc finger protein 292 |
| chr7_-_127392687 | 0.15 |
ENST00000393313.5
ENST00000619291.4 ENST00000265827.8 ENST00000434602.5 |
ZNF800
|
zinc finger protein 800 |
| chr7_-_14903319 | 0.15 |
ENST00000403951.6
|
DGKB
|
diacylglycerol kinase beta |
| chr11_+_73308237 | 0.15 |
ENST00000263674.4
|
ARHGEF17
|
Rho guanine nucleotide exchange factor 17 |
| chr9_-_120714457 | 0.15 |
ENST00000373930.4
|
MEGF9
|
multiple EGF like domains 9 |
| chr7_-_28180735 | 0.15 |
ENST00000283928.10
|
JAZF1
|
JAZF zinc finger 1 |
| chr9_+_127612257 | 0.14 |
ENST00000637173.2
ENST00000630492.2 ENST00000627871.2 ENST00000373302.8 ENST00000373299.5 ENST00000650920.1 ENST00000476182.3 |
STXBP1
|
syntaxin binding protein 1 |
| chr6_-_70957029 | 0.14 |
ENST00000230053.11
|
B3GAT2
|
beta-1,3-glucuronyltransferase 2 |
| chr8_+_63168597 | 0.14 |
ENST00000539294.6
ENST00000621957.4 ENST00000517371.5 ENST00000621413.4 ENST00000612880.4 |
YTHDF3
|
YTH N6-methyladenosine RNA binding protein 3 |
| chr10_+_92291155 | 0.14 |
ENST00000358935.3
|
MARCHF5
|
membrane associated ring-CH-type finger 5 |
| chr2_+_46698909 | 0.14 |
ENST00000650611.1
ENST00000306503.5 |
LINC01118
SOCS5
|
long intergenic non-protein coding RNA 1118 suppressor of cytokine signaling 5 |
| chr8_-_123396412 | 0.14 |
ENST00000287394.10
|
ATAD2
|
ATPase family AAA domain containing 2 |
| chr10_+_110871903 | 0.14 |
ENST00000280154.12
|
PDCD4
|
programmed cell death 4 |
| chr1_+_244835616 | 0.14 |
ENST00000366528.3
ENST00000411948.7 |
COX20
|
cytochrome c oxidase assembly factor COX20 |
| chr1_+_61082553 | 0.13 |
ENST00000403491.8
ENST00000371187.7 |
NFIA
|
nuclear factor I A |
| chr19_-_49441508 | 0.13 |
ENST00000221485.8
|
SLC17A7
|
solute carrier family 17 member 7 |
| chr12_-_54385727 | 0.13 |
ENST00000551109.5
ENST00000546970.5 |
ZNF385A
|
zinc finger protein 385A |
| chr2_+_199911285 | 0.13 |
ENST00000319974.6
|
C2orf69
|
chromosome 2 open reading frame 69 |
| chr1_-_32702736 | 0.13 |
ENST00000373484.4
ENST00000409190.8 |
SYNC
|
syncoilin, intermediate filament protein |
| chrX_+_70444855 | 0.13 |
ENST00000194900.8
|
DLG3
|
discs large MAGUK scaffold protein 3 |
| chr7_+_139341311 | 0.13 |
ENST00000297534.7
ENST00000541515.3 |
FMC1
FMC1-LUC7L2
|
formation of mitochondrial complex V assembly factor 1 homolog FMC1-LUC7L2 readthrough |
| chr1_+_200739542 | 0.12 |
ENST00000358823.6
|
CAMSAP2
|
calmodulin regulated spectrin associated protein family member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of AGUGCUU
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0072034 | primary prostatic bud elongation(GO:0060516) renal vesicle induction(GO:0072034) |
| 0.2 | 1.0 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.2 | 1.4 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.2 | 1.9 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 0.4 | GO:1903248 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.1 | 0.4 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.1 | 0.6 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.1 | 0.6 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.1 | 0.6 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.1 | 0.6 | GO:0061290 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.1 | 0.3 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.1 | 1.4 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.1 | 0.5 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.1 | 1.2 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 0.3 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.1 | 0.3 | GO:0061642 | chemoattraction of serotonergic neuron axon(GO:0036517) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) chemoattraction of axon(GO:0061642) negative regulation of cell proliferation in midbrain(GO:1904934) planar cell polarity pathway involved in midbrain dopaminergic neuron differentiation(GO:1904955) |
| 0.1 | 0.5 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.1 | 1.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.2 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.1 | 0.2 | GO:1903998 | regulation of eating behavior(GO:1903998) |
| 0.1 | 0.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 1.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.4 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.2 | GO:1903892 | negative regulation of ATF6-mediated unfolded protein response(GO:1903892) |
| 0.1 | 0.2 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.1 | 0.4 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 1.1 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.3 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.0 | 0.2 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.0 | 0.5 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.2 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.2 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.0 | 0.4 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.2 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.6 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.3 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.1 | GO:2000559 | CD24 biosynthetic process(GO:0035724) activation of meiosis involved in egg activation(GO:0060466) negative regulation of monocyte extravasation(GO:2000438) regulation of CD24 biosynthetic process(GO:2000559) positive regulation of CD24 biosynthetic process(GO:2000560) |
| 0.0 | 0.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.0 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.2 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.3 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.2 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.4 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.2 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.5 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.0 | 0.2 | GO:0061083 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 2.2 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.4 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.4 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.3 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.0 | 0.2 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:1903294 | regulation of glutamate secretion, neurotransmission(GO:1903294) positive regulation of glutamate secretion, neurotransmission(GO:1903296) |
| 0.0 | 0.1 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) platelet alpha granule organization(GO:0070889) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.2 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 1.1 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.3 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.2 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.3 | GO:0031577 | spindle checkpoint(GO:0031577) |
| 0.0 | 0.5 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.5 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.2 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.6 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.3 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 1.0 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 0.3 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.9 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.0 | 0.0 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.4 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.5 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.5 | GO:0008209 | androgen metabolic process(GO:0008209) |
| 0.0 | 0.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.3 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.8 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.2 | 0.6 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.1 | 1.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.6 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 1.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.5 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 0.2 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 1.4 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.1 | GO:0060203 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.0 | 0.8 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 1.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.2 | 1.3 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 0.4 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.1 | 0.4 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.1 | 0.4 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.1 | 0.3 | GO:0005115 | receptor tyrosine kinase-like orphan receptor binding(GO:0005115) chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.1 | 0.3 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 2.1 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 2.4 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 0.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 0.6 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.2 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.1 | 0.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.4 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.3 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 1.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.3 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.5 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.3 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0036505 | prosaposin receptor activity(GO:0036505) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.7 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 1.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.5 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.6 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.3 | GO:0071532 | oxygen sensor activity(GO:0019826) ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.2 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.2 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.0 | 0.3 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.6 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.2 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 1.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.5 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.9 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 2.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.2 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.6 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.4 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |